First Steps for the Reproducible Research Novice
http://www.riffomonas.org/reproducible_research/first_steps/ Press 'h' to open the help menu for interacting with the slides
Welcome back to the Riffomonas Reproducible Research Tutorial Series. Today's tutorial is called "First Steps for the Reproducible Research Novice." The approach is really inspired by my interactions with the writings of Christie Bahlai and Karl Broman, two scientists that I really respect for their practical and down-to-earth advice when it comes to improving the reproducibility of data analysis workflows.
In today's tutorial, we'll go over some first practical steps to improving the reproducibility of our data analyses. As you hopefully recall from the previous tutorials, we read an editorial called All Hail Reproducibility by Jacques Ravel and Eric Wommack. In that, they mentioned several tools. In this tutorial, we're going to use several of these tools by setting up ORCID and GitHub accounts, exploring FigShare, and learning more about Markdown.
Finally, we'll do a simple exercise using GitHub and Markdown that will be the useful analogy for thinking about the reproducibility of larger projects. Join me in opening the slides for this tutorial, which you can find as the "First Steps Towards Reproducibility" tutorial within the Reproducible Research Tutorial Series at riffomonas.org.
So, let's dig into this tutorial, the "First Steps for the Reproducible Research Novice."
Learning goals
- Conclude that reproducible research is a continual process with no end
- Evaluate initial steps you can take to make your research more reproducible
- Explore interfaces that can make your research more reproducible
- Write a simple document in markdown
- Use version control without knowing it
The learning goals for this tutorial include reaching the conclusion that reproducible research is a continual process with no end. We'll evaluate the initial steps that you can take to make your research more reproducible. We'll explore different interfaces that you can use to make your research more reproducible.
Then we'll learn a very useful tool called Markdown that allows us to write simple or even more complex documents, and then finally, we'll use a tool called version control without even knowing it.
Baby steps
credit: Christie Bahlai and Karl Broman
- Don't feel like you need to make everything 100% reproducible in your first go
As I mentioned in the introduction, a lot of my thinking in how we've structured these tutorials comes from Christie Bahlai and Karl Broman, two data analyst people that I really respect for their clear thinking and practical advice.
And so, if you click on the links in the bottom-left corner, those links will take you to different blog posts that they've written about reproducible research and the idea of the "First Steps for the Reproducible Research Novice." So, the first point I want you to keep in mind is, don't feel like you need to make everything 100% reproducible in your first go.
That's a huge goal, and we might like to be smug and think all research should be reproducible, but as we've already discussed in the previous tutorials, that's a high bar to try to reach. Second, you'll quickly realize, as I've already mentioned, that that's a fool's errand.
Baby steps
credit: Christie Bahlai and Karl Broman
Don't feel like you need to make everything 100% reproducible in your first go
You will quickly realize that is a fool's errand - nothing will ever be fully reproducible
As I mentioned in the introduction, a lot of my thinking in how we've structured these tutorials comes from Christie Bahlai and Karl Broman, two data analyst people that I really respect for their clear thinking and practical advice.
And so, if you click on the links in the bottom-left corner, those links will take you to different blog posts that they've written about reproducible research and the idea of the "First Steps for the Reproducible Research Novice." So, the first point I want you to keep in mind is, don't feel like you need to make everything 100% reproducible in your first go.
That's a huge goal, and we might like to be smug and think all research should be reproducible, but as we've already discussed in the previous tutorials, that's a high bar to try to reach. Second, you'll quickly realize, as I've already mentioned, that that's a fool's errand.
Nothing, nothing will ever be fully reproducible. We can do our best, but over the long arc of time, things change, right? We've mentioned this in the previous tutorials. Databases change, software change, there are differences between different operating systems, and so nothing will ever be fully, 100% reproducible. What I encourage people to do is, with each project, attempt to add one small element to make your work more reproducible.
Baby steps
credit: Christie Bahlai and Karl Broman
Don't feel like you need to make everything 100% reproducible in your first go
You will quickly realize that is a fool's errand - nothing will ever be fully reproducible
With each project, attempt to add one small element to make your work more reproducible
As I mentioned in the introduction, a lot of my thinking in how we've structured these tutorials comes from Christie Bahlai and Karl Broman, two data analyst people that I really respect for their clear thinking and practical advice.
And so, if you click on the links in the bottom-left corner, those links will take you to different blog posts that they've written about reproducible research and the idea of the "First Steps for the Reproducible Research Novice." So, the first point I want you to keep in mind is, don't feel like you need to make everything 100% reproducible in your first go.
That's a huge goal, and we might like to be smug and think all research should be reproducible, but as we've already discussed in the previous tutorials, that's a high bar to try to reach. Second, you'll quickly realize, as I've already mentioned, that that's a fool's errand.
Nothing, nothing will ever be fully reproducible. We can do our best, but over the long arc of time, things change, right? We've mentioned this in the previous tutorials. Databases change, software change, there are differences between different operating systems, and so nothing will ever be fully, 100% reproducible. What I encourage people to do is, with each project, attempt to add one small element to make your work more reproducible.
So, with this project, identify something that we do here. Perhaps it's setting up a FigShare account and making sure that all of your derivative data that goes into making a plot ends up on FigShare. Perhaps it's being really diligent about putting your sequence data and your metadata into the Sequence Read Archive. So, with each project, you layer on new skills, new elements to make your research more reproducible.
First steps to reproducibility
ORCID
Post data behind your figures
Making your code publicly available
If you get nothing else out of this workshop, these three steps will significantly enhance your ability to perform reproducible science
ORCID
- Why:
- Will your current email address be any good in 10 years?
- Ties in other services (e.g. ImpactStory, LinkedIn, ResearchGate)
- It's free!
- How:
So, some first steps that I've already mentioned, you could get an ORCID account. So many journals nowadays require the corresponding author at least to have an ORCID account. You can post the data behind your figures into a tool like FigShare.
You can make your code publicly available, and then also, I didn't list it here because it's kind of…I assume it in people, but make sure that your sequence data is publicly available in a database, and the appropriate database for sequence data is the Sequence Read Archive, the SRA. So, the first tool that we're going to talk about is ORCID.
And so, to motivate this, ask yourself a question. If you publish a paper and your email address is on that manuscript, or you put out some tool or some resource that's attached to your email address, do you think your current email address will be any good in 10 years? Right?
I'd like to think that my email address here at the University of Michigan will still be valid in 10 years, but I know that 10 years ago, I did not have a umich.edu email address. I was at the University of Massachusetts thinking, well, I'll be there for 10 years, right? So things change. Sure, there's Google to get a hold of people. Pat Schloss is kind of a unique name, but if your name is Joe Smith or Mary Smith, those are kind of hard names to Google, and so it's difficult to link back to people.
And so, the ORCID ID is a tool that allows you to connect your scholarly work with your identity, right? And so, it helps to deal with changing email addresses, changing contact information, as well as the problem that some of us have names that are kind of common, right? And so, the other advantage is that it ties into other services, so if you're using a service like ImpactStory, LinkedIn, or ResearchGate, they'll allow you to populate information there using your ORCID IDs.

It's also free, which is really nice, and unlike these other services like LinkedIn, ORCID doesn't send you spam. At least, I haven't noticed any great amounts of spam. So, let's go ahead and click on this link for ORCID ID, and this brings up the ORCID ID homepage, okay?
And so, as they say, it allows you to "distinguish yourself in three easy steps," and that the ORCID ID provides this "persistent digital identifier" that allows you to "distinguish yourself from every other researcher," okay? And it's being used in manuscript and grant submission, it helps support automated linkages between you and your professional activities, making sure that your work is recognized.
So, what I'd like you to do is go ahead and register to "Get your own unique ORCID identifier." So, we'll go ahead and click on "Register now!" I already have an account, but go ahead and plug in your first name, your last name, your primary email, your additional email, and then go ahead and create your ORCID password, and you can tell them how often you want to be bugged, and go ahead and register.
So, I'm going to go ahead and sign into my account. And I've been on ORCID for a while, and this is something that might require a fair amount of curation, depending on what you want to provide.
I know we certainly get asked a lot of places for information that makes something like an electronic CV. I think ORCID is probably something worth investing a fair amount of time in. But you can see that it has a number of my publications in here.
I notice there's a few missing that I should probably go ahead and add. It also has information here asking about education, employment, funding sources, keywords, various bits of information. And so, again, this is a site that someone could come to to learn about me, okay? To illustrate this, if we go to PubMed…and let's look for one of my papers.
And…Here's a editorial that I wrote in the journal mBio that we'll go ahead and click on, and this will take me to the mBio website here.
And if you look down here under ORCID, on the left…on the right side of the screen, sorry, you'll see there's a profile for Schloss, P. D., profile for Casadevall, A., Arturo Casadevall. So, let's go ahead and see what Arturo already has in here. And so, he doesn't have any information that is publicly available, which is fine. If we come back and we look at my profile, you'll see that I… This is my public profile, right?
So this is what you would see about me if you came and looked at my information, okay? And so, you could see all the papers that I've written. And I can imagine that, like I said, this would be useful for people that have a name that's fairly common, that's not unique within science, for people that perhaps move around a lot, for people who change their name. I know several women who have changed their name over the course of their scientific career.
But again, it's a useful tool for keeping track of your identity with your scholarly work. And you can see, Arturo's had very little information in it. Mine has a bit of information, right? It links you to my works and I could add more information if I wanted to. Ultimately, the value of ORCID comes from what do you want to get out of it, okay?
And so, the first thing I'd encourage you to do is to start populating this with information. It is useful. I also know that we get inundated by a lot of requests to create things like this and other useful tools like your Google Scholar profile, but there, again, that's one more thing to do.
The advantage of ORCID is that it's nonprofit, you control it. The advantage of something like Google Scholar is that everybody uses Google, but at the same time, it's commercial and you're not quite sure what's going on with the information. And so, ORCID is, again, a tool that many journals are requiring at least one of the authors, typically the corresponding author, have an ORCID ID. So, if for no other reason, it makes sense to have an ORCID ID.
Post data behind your figures
- What's wrong with posting data on a private server?
So, returning to the slides, now I want to talk briefly about posting the data behind your figures. This is something really simple that you can do to make your work more reproducible and more transparent. So, one question might be, "Well, what's wrong with posting the data on a private server? My lab has a website. Why don't I just put all the data for my lab on that website?"
So, think to yourself, "Well, what could be a problem with that?" And so, one problem would be that, again, you might move. You might leave science. Your PI that you work for now might move. My PI while I was a postdoc moved institutions. My graduate adviser retired in the last few years.
So, if we'd have put data on their servers, it would've been difficult to track down that data. In addition, other problems with it is that there's no real standards being enforced on these private servers. One of the advantages of posting sequence data to the Sequence Read Archive is that they have a standard called MIMARKS that specifies the type, the minimum amount of information that must be provided to describe your sequence data, okay?
So, I could post data on my own website. To be in full disclosure, I have done that. It's not something I'm proud of. And we did post MIMARKS data, but at the same time, there's nothing requiring us to do that. And so, yeah, you can post your sequence data to your lab website, but there's no standards.
Post data behind your figures
- What's wrong with posting data on a private server?
- Sequence data and metadata must be posted to SRA
So, returning to the slides, now I want to talk briefly about posting the data behind your figures. This is something really simple that you can do to make your work more reproducible and more transparent. So, one question might be, "Well, what's wrong with posting the data on a private server? My lab has a website. Why don't I just put all the data for my lab on that website?"
So, think to yourself, "Well, what could be a problem with that?" And so, one problem would be that, again, you might move. You might leave science. Your PI that you work for now might move. My PI while I was a postdoc moved institutions. My graduate adviser retired in the last few years.
So, if we'd have put data on their servers, it would've been difficult to track down that data. In addition, other problems with it is that there's no real standards being enforced on these private servers. One of the advantages of posting sequence data to the Sequence Read Archive is that they have a standard called MIMARKS that specifies the type, the minimum amount of information that must be provided to describe your sequence data, okay?
So, I could post data on my own website. To be in full disclosure, I have done that. It's not something I'm proud of. And we did post MIMARKS data, but at the same time, there's nothing requiring us to do that. And so, yeah, you can post your sequence data to your lab website, but there's no standards.
There's no enforcement as to the quality of the data, and so if you post to a third party website like the Sequence Read Archive, there's certain requirements and there's certain rubrics that you need to follow so that your data can be posted there.
Post data behind your figures
- What's wrong with posting data on a private server?
- Sequence data and metadata must be posted to SRA
- Other data should be posted to stable web server (e.g. FigShare)
- Provides DOIs for each set of data
- Prohibits user from deleting data
- Is free (within limits)
So, returning to the slides, now I want to talk briefly about posting the data behind your figures. This is something really simple that you can do to make your work more reproducible and more transparent. So, one question might be, "Well, what's wrong with posting the data on a private server? My lab has a website. Why don't I just put all the data for my lab on that website?"
So, think to yourself, "Well, what could be a problem with that?" And so, one problem would be that, again, you might move. You might leave science. Your PI that you work for now might move. My PI while I was a postdoc moved institutions. My graduate adviser retired in the last few years.
So, if we'd have put data on their servers, it would've been difficult to track down that data. In addition, other problems with it is that there's no real standards being enforced on these private servers. One of the advantages of posting sequence data to the Sequence Read Archive is that they have a standard called MIMARKS that specifies the type, the minimum amount of information that must be provided to describe your sequence data, okay?
So, I could post data on my own website. To be in full disclosure, I have done that. It's not something I'm proud of. And we did post MIMARKS data, but at the same time, there's nothing requiring us to do that. And so, yeah, you can post your sequence data to your lab website, but there's no standards.
There's no enforcement as to the quality of the data, and so if you post to a third party website like the Sequence Read Archive, there's certain requirements and there's certain rubrics that you need to follow so that your data can be posted there.
Other options for, say, non-sequence data would be to post your data to a stable website. So, one of the problems with posting to your lab server is, that lab server might move, the address might change, the PI might leave, you might leave, and so, posting to a stable third party like FigShare is a useful tool for making those data publicly available, okay? And so, the advantage of something like FigShare is that it provides a DOI, which is a digital object identifier for each set of data.
And so, this is a persistent identifier that has a web link, a web address, that goes directly to your dataset. They also prohibit users from deleting data, all right? So, once you put it onto FigShare and you make it public, it's public. It cannot be deleted. And then, within limits of file sizes, it's also free.
So, something you might be saying in the back of your head is, "Well, why don't I just submit this data as supplementary material for my publication?" And you could do that, right? And again, I've done that, as well. The challenge with that is that oftentimes, a journal, when they publish supplementary material, they convert it to weird formats, all right?
So, I know people that have gotten sequence data out of supplementary material that's in a PDF. That's not going to be useful for any downstream analysis. Whereas if something's available in a FigShare, you can post it as a FASTA-formatted file. Also, it would be great if all data was available from all published papers or that all data got published, but the fact is that not all data gets published, and we might like to have a way to access data from unpublished studies.
There might be studies that are never published, and so, using a tool like FigShare makes sure that our data gets out there, and that enables other people to use. And again, because it has a DOI, people can then cite, and should cite, where they got our data from, okay? So, an activity that we're going to do now, is, we're going to go into FigShare, and we're going to try to find the data that was described in the Wommack and Ravel editorial from James Meadow and colleagues, okay?
Post data behind your figures
- What's wrong with posting data on a private server?
- Sequence data and metadata must be posted to SRA
- Other data should be posted to stable web server (e.g. FigShare)
- Provides DOIs for each set of data
- Prohibits user from deleting data
- Is free (within limits)
- Can you find the FigShare accession from Meadow et al?
So, returning to the slides, now I want to talk briefly about posting the data behind your figures. This is something really simple that you can do to make your work more reproducible and more transparent. So, one question might be, "Well, what's wrong with posting the data on a private server? My lab has a website. Why don't I just put all the data for my lab on that website?"
So, think to yourself, "Well, what could be a problem with that?" And so, one problem would be that, again, you might move. You might leave science. Your PI that you work for now might move. My PI while I was a postdoc moved institutions. My graduate adviser retired in the last few years.
So, if we'd have put data on their servers, it would've been difficult to track down that data. In addition, other problems with it is that there's no real standards being enforced on these private servers. One of the advantages of posting sequence data to the Sequence Read Archive is that they have a standard called MIMARKS that specifies the type, the minimum amount of information that must be provided to describe your sequence data, okay?
So, I could post data on my own website. To be in full disclosure, I have done that. It's not something I'm proud of. And we did post MIMARKS data, but at the same time, there's nothing requiring us to do that. And so, yeah, you can post your sequence data to your lab website, but there's no standards.
There's no enforcement as to the quality of the data, and so if you post to a third party website like the Sequence Read Archive, there's certain requirements and there's certain rubrics that you need to follow so that your data can be posted there.
Other options for, say, non-sequence data would be to post your data to a stable website. So, one of the problems with posting to your lab server is, that lab server might move, the address might change, the PI might leave, you might leave, and so, posting to a stable third party like FigShare is a useful tool for making those data publicly available, okay? And so, the advantage of something like FigShare is that it provides a DOI, which is a digital object identifier for each set of data.
And so, this is a persistent identifier that has a web link, a web address, that goes directly to your dataset. They also prohibit users from deleting data, all right? So, once you put it onto FigShare and you make it public, it's public. It cannot be deleted. And then, within limits of file sizes, it's also free.
So, something you might be saying in the back of your head is, "Well, why don't I just submit this data as supplementary material for my publication?" And you could do that, right? And again, I've done that, as well. The challenge with that is that oftentimes, a journal, when they publish supplementary material, they convert it to weird formats, all right?
So, I know people that have gotten sequence data out of supplementary material that's in a PDF. That's not going to be useful for any downstream analysis. Whereas if something's available in a FigShare, you can post it as a FASTA-formatted file. Also, it would be great if all data was available from all published papers or that all data got published, but the fact is that not all data gets published, and we might like to have a way to access data from unpublished studies.
There might be studies that are never published, and so, using a tool like FigShare makes sure that our data gets out there, and that enables other people to use. And again, because it has a DOI, people can then cite, and should cite, where they got our data from, okay? So, an activity that we're going to do now, is, we're going to go into FigShare, and we're going to try to find the data that was described in the Wommack and Ravel editorial from James Meadow and colleagues, okay?
So, I'm going to try to find the Meadow et al. article in PubMed, and I will then go, Meadow J [au], and that was published in the journal Microbiome.
And so, this should get us pretty close. And yes, there's two articles that Meadow wrote in the journal Microbiome. This is the one we want, and I'll go ahead and click on the Microbiome journal version.
And what I'm looking for now is mention of the FigShare data, and so, hopefully, that's going to be provided in the Methods section here, so I'm kind of scanning this. I'm going to get rid of this popup. Go away. Ah-ha. And so, you see, "Sequence files and metadata for all samples used in this study have been deposited in Figshare," okay?
So, this was one of the things we critiqued their GitHub repository for, their paper for, was that, yes, they make their sequence files and metadata publicly available, but it's in FigShare. It's not in the Sequence Read Archive, and that's…it's great that the data are available.
We'd like it to be in the Sequence Read Archive, again, because the SRA is going to impose certain standards on what type of metadata are provided. So, we can go ahead and click on this link, and this then brings us to the Lillis_Classroom_Surfaces page at FigShare. We can see information about how to cite it.
We can download it. It's about 200 megabytes, and it's tagged with different keywords. It's got a description of what's going on. They tell you how to cite it and where it's published. We see that there's a swab.seqs.fna as well as the metadata file. So, one of the things I noticed is that this is an FNA file, which tells me that this is the A, Ts, Gs, and Cs.
It's not going to include for me the quality scores from the MiSeq sequence data. And so, that's, again, one of the problems with posting it to FigShare rather than the SRA is that the SRA probably would've required them to also post the quality score data, and that quality score data is now useful four or five years later for people that want to come back and reanalyze the data using more modern methods of denoising sequences.
Also, I see that these probably aren't the raw data, because it's a single FASTA file rather than the pairs. If they did paired-end sequencing of the 16S fragments, there should be two files, but there's only one here, okay? So, they've given us the 16S data, but it's not exactly the raw data, okay? So, again, it's… Don't get me wrong, it's great that the data are available on FigShare, but it would've been ideal for them to have been posted on the Sequence Read Archive.
To absolve them a little bit, back when they submitted this, probably the Sequence Archive was really difficult to work with in getting their data into the website. Through tools like those available in the Mothur software package, it's now considerably easier to post your data into the Sequence Read Archive, and that's really what you should be doing, okay?
But again, this is illustrative of the type of data, or of data that you can then post to FigShare. You might think of any figure that you have in your paper, the data that you're plotting, you could make a file, you can make an accession like this, and you could post the data that's in that figure onto FigShare.
So, say you've got a plot showing the diversity of different samples in people with healthy colons versus those with adenomas or carcinomas, you could post the spreadsheet file to FigShare that was used…you could post that spreadsheet that was used to make the plot here to FigShare that others then could then take the data and do whatever they want with it.
And again, this is another somewhat simple thing that you could do to increase the reproducibility of your data, so not only make your sequence data available at the SRA, but you could also post the data behind your figures. And as Meadow did, be sure that you also provide a link or the DOI to the FigShare accession in your paper. Typically, that would go in the Methods section.
Making your code publicly available
So, the next thing that you can do to improve the reproducibility of your research is to make your code publicly available. Many of us have no doubt had that problem where you read a paper and you see, "Custom scripts were used, available upon request from the PI." Well, as we've already mentioned, people move, people are hard to get ahold of.
Sometimes that code isn't in really good shape. And so, a tool that we can use to make our code publicly available, and it could be code, or methods, descriptions, or anything like that, is a website called github.com. And so, what I'd like to do now is go with you into GitHub.
And I'm going to log out of my account. And so, you should get a webpage that looks like this, unless you already have a GitHub account, okay? And so, it's a pretty attractive site. They kind of tell you a bit about GitHub, but what I want you to do right now is to go ahead and pick a username, enter your email address as well as a password, and go ahead and click "Sign up for GitHub."
One thing that some people try to do is that it's easy to get many usernames here, right? So you might have a username for your email, you might have a username for your Twitter account, you might have a username for GitHub. Sometimes it's nice to have a common username across sites, across platforms. That's not always possible, but try.
Making your code publicly available
Once you do that, once you sign up for GitHub, it allows you to then sign in. I'm going to go ahead and get rid of that. I'm going to go ahead and sign in to my GitHub account. And this is a profile from my GitHub account which, if you're just signing up, yours will look much different than this.
And so, this is kind of the profile of somebody that's fairly active in using GitHub for a lot of their research. I'm going to go ahead and click in the upper-right corner here to view Your Profile. And so, you can add a picture. This is our lovely family pig, my name.
You'll see I've put my ORCID ID into GitHub. My affiliation, contact information. And if you're just starting on GitHub, you won't have any repositories. You won't have any contributions, okay? But over the course of this tutorial series, you'll start to create repositories and you'll start to have more information here, okay?
So, we're going to go ahead now and create a repository. So, we're going to click up here on this plus sign and we're going to click "New repository" and the repository name we're going to use is rr_practice.
Making your code publicly available
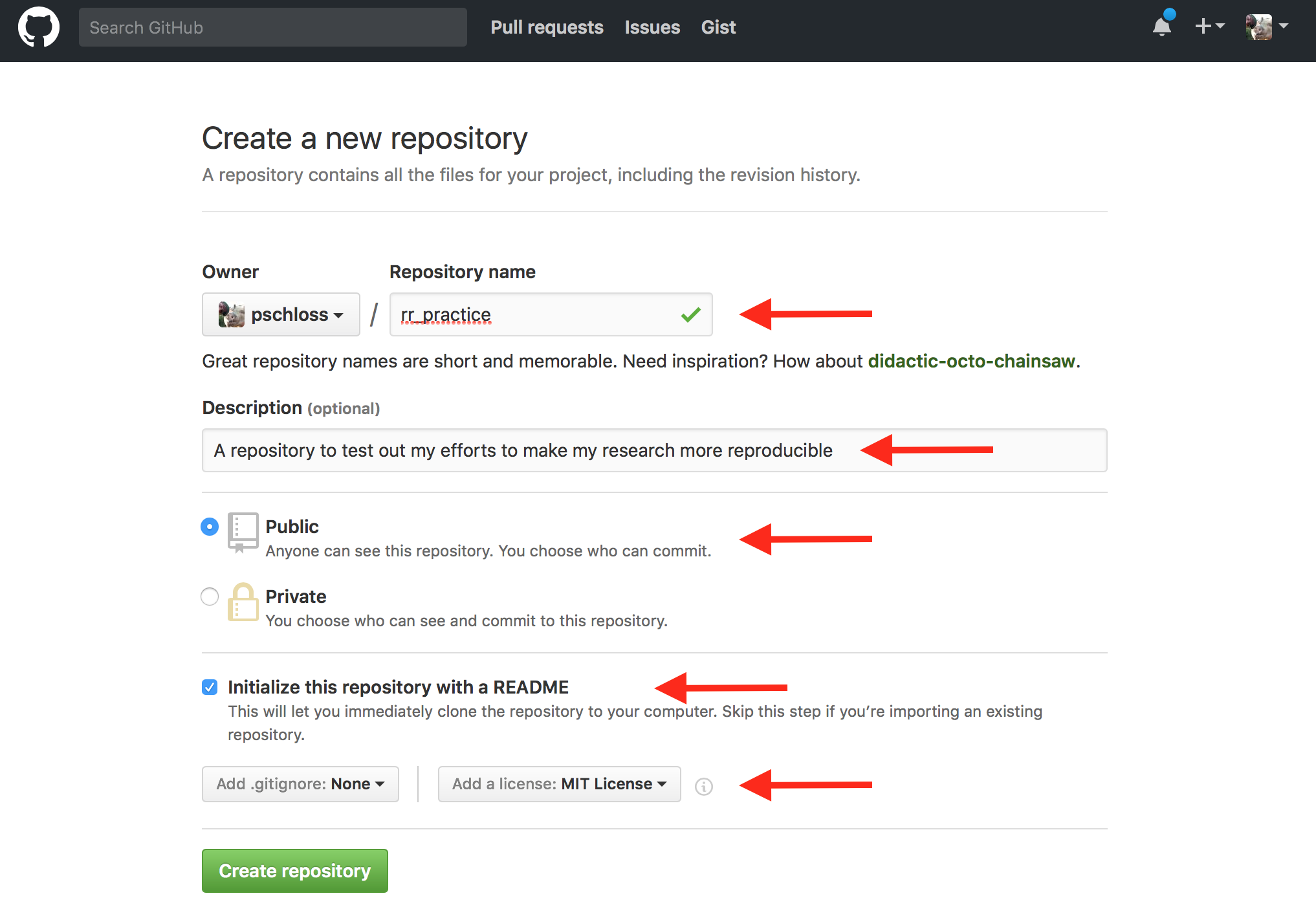
So, reproducible research practice. I think if you wanted to use a space...it's going to tell you that it's going to create it as rr_practice.
So, spaces are bad news when it comes to doing a lot of bioinformatics research, so I'm going to call mine rr_practice. For a description, which is optional, I'm going to say, "A repository to test out my efforts and to make my research more reproducible." You can put whatever description you want.
For whatever reason, that's what I came up with. I'm going to make mine public. You should, too. GitHub allows you to have unlimited public repositories for free, but you'd have to pay if you want to use a private repository. That being said, if you have an academic account, you can use GitHub with an academic account and get permission from them or a setting on your account to have as many private accounts as you want.
Just be sure that by the time you submit your manuscript that you switch your repository from being private to public. But for this exercise and for the purposes of this tutorial series, everything should be done in the public, okay? We're going to then also go ahead and click "Initialize this repository with a README," and for now, I'm going to not add a .gitignore file, and I'm going to add a license, the MIT License, okay?
Making your code publicly available
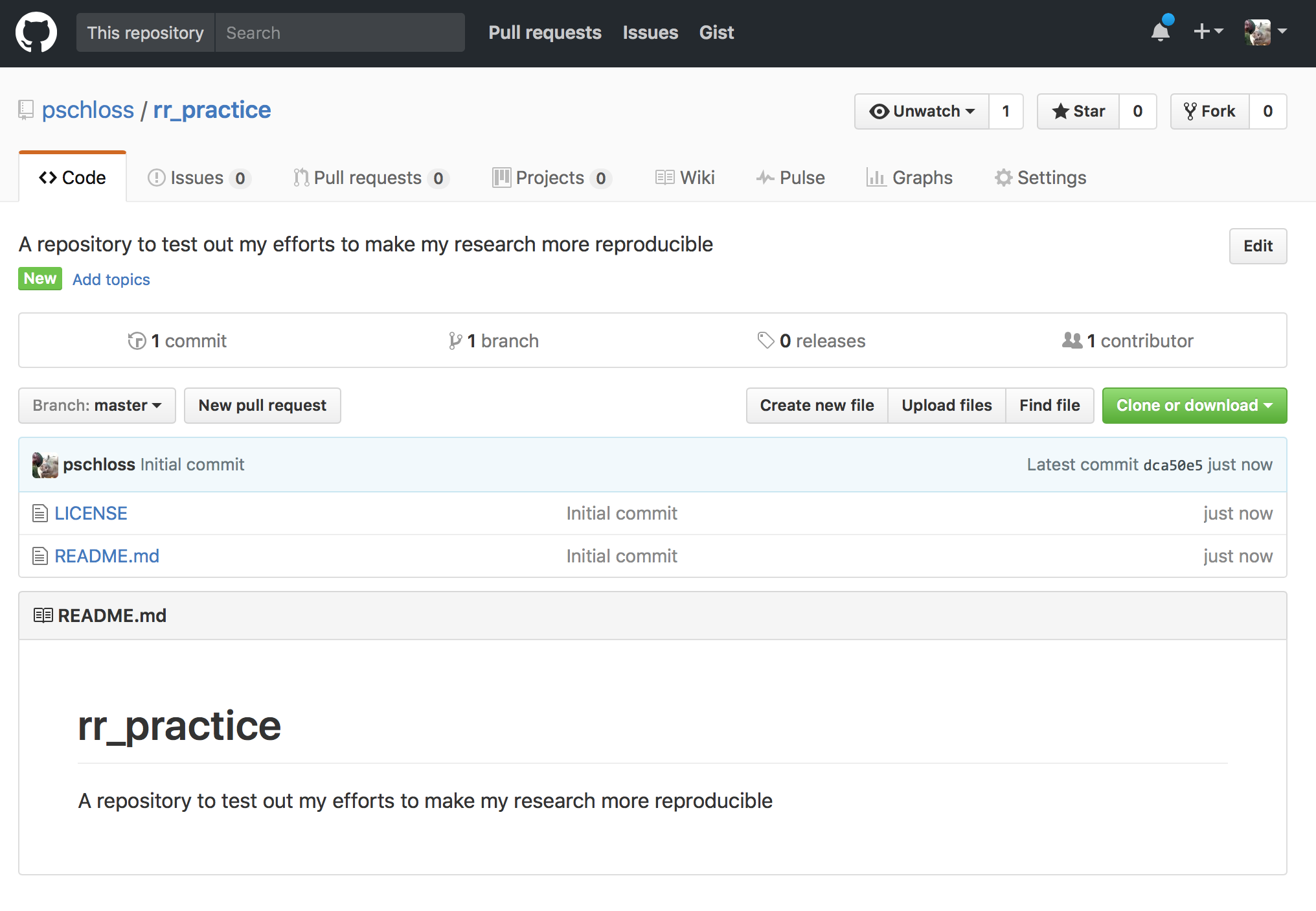
So, it should look something like this, and we're going to go ahead and click this wonderful green button. It's very exciting, creating your first repository. And voilà, here we go. Your first repository. You might recall what the Meadow et al. repository looked like, and it looked vaguely like this. They had a few more files and I think a directory or two in there.
And so, now, you have a license and you have a file called README.md, okay? So, go ahead and click on this blue README link and this brings you to the README file as it's been rendered.
Making your code publicly available
It's got a title, rr_practice. "A repository to test out my efforts to make my research more reproducible." And so, go ahead and click on the pencil, which gives you a balloon that says "Edit this file," and we see that this now looks like a text file. I can click in here and I get a flashing cursor, okay?
So, you'll see this file is called README.md. The MD is a shorthand that tells you that this is a Markdown-formatted file, okay? And so, that's different than, say, a DOCX file which would be a Word file. A DOCX Word file is a binary-formatted file that's somewhat proprietary.
Making your code publicly available
- A file ending in
mdindicates that it is a file in "markdown" format - This is a text file (not a Word file)
- There are simple "markups" that you can do to add formatting to the text
An MD file is a text file that has no built-in formatting, nothing, no special sauce. It's a plain text file. And so, what you perhaps noticed was that previously, the title of this file within the document was rr_practice. So, we can go ahead and click "Preview change," and we see that there's some formatting created here, right?
So, we can toggle back and forth, and we can see that some formatting is applied, and you see a pound sign here to create that heading. And there are various tutorials that you can see online for formatting Markdown. It's meant to be pretty simple. I'm going to go over a handful of simple things with you.
Making your code publicly available
- You can explore GitHub-flavored markdown further
- Make the following changes and toggle back and forth from the preview...
#to##or###effortsto*efforts*,**efforts**,***efforts***, or~~efforts~~- Add a bulleted list
Type along with me. I'm going to put, "This is a heading," after two pound signs, okay? If I do a third one, "This is another heading," okay? And here, I'm going to put, "Here is some text to describe the overall goal of my data analysis plan," okay?
And if I come down, I can say, "Here is some more text describing how to use markdown." Okay? So, we've created various headings, and if we look at them here, we now see that we get different-sized headings depending on how many pound signs we used.
These green bars in the margin is GitHub's way of telling us that we've added this information to the file. If we come back here, we've seen that we can change heading sizes with the number of pound signs, okay? So, you can look at this text file and get a sense of the organization, all right?
So, I could add a third heading, say… Or let's make it look a bit more like a paper. So, if I say this is Introduction. We can come down and say, "Results, Discussion, Methods," all right?
And so, perhaps I'll delete this and clean this up a bit and say, "This is…" Or our Methods would be…let's say, "Experimental design." Down here I might say, "DNA extraction and amplification," all right?
And you could keep going. And in here, I could say, so, "We obtained 200 samples from various soils across the state of Michigan." Okay? "We used a bead-beating DNA extraction kit." Okay? "We amplified the 16S rRNA gene."
Perhaps I say, "We amplified the V4 region of the 16s rRNA gene." Okay? So, we added some text. This looks vaguely like a paper or a lab report. We can then preview the changes and see that this is starting to take shape, okay?
So, those are headings, allows you to structure the information. Something else we could do is that we can…this isn't typically done in a paper, but we can add different types of emphasis, okay? So, we might say…let's put a single star. So, we add a star before "repository" and we see that GitHub has nicely started to format "repository" for us to be italicized.
And so, if you add another star at the end, we see the rest of the text goes back to being a normal font. So, if we look at this, we now see "repository" has been emphasized, has been made italics, okay? So, let's put "my efforts"after two stars, okay?
And so, you'll see already that GitHub, in their interface here, has made that bold. And so, here, again, GitHub is doing some other magic under the scenes where it's…the strikethrough is showing that it's deleting "my efforts" and it's adding "my efforts" in bold.
And so, down here, maybe I'll do three stars, and as we can see from the magic sauce that GitHub is using, "describe" now is going to be bolded and italicized. And sure enough, "describe" is bolded and italicized, okay? Another useful tool that I like to do when outlining is to perhaps make a bulleted list.
And so, here in my Results section, I'm going to "Describe the site that I obtained samples from," okay? And so, you'll see that I'm using these asterisks on the first line to create a bulleted list. I'll then say, "Differences in richness between types of sites."
Then I'll say, "Differences in beta-diversity between types of sites," okay? This is not a super-exciting paper, but I just want to illustrate the point that you can use these stars here in the text mode, we can visually see that's a bulleted list. And we can also then preview the changes and see that on the website, at least, GitHub will format that to be a bulleted list, okay?
So, this will get you far, using the stars, and the pound signs, and the bulleted lists, but there's other things that you might want to do, like adding hyperlinks, or pictures, or blockquotes and things like that. So, how do we figure out what GitHub uses and what Markdown uses to utilize those features? So, let's go to our friend Google, and I'm going to use "github flavored markdown" and search for that.
And what we want…this link is going to be the helpful one, "Mastering Markdown GitHub Guides." And here, you'll see a page where they have different examples. We've already talked about different types of headers, emphasis, types of lists, images, links, things like that, okay?
Exercise
- Edit the README.md file
- Give the file a more descriptive title using a
#header - Write instructions for how to fold a paper airplane
- Give the file a more descriptive title using a
- Look at the bottom of the README.md page
- In the narrow box below "Commit changes" enter a brief description of what you changed in the file
- In the larger box, write a more detailed description
- Press the green "Commit changes" box
So...what I'd like you to do next is, within this README file, to go ahead and delete everything and create a title that says, "How to fold a paper airplane," okay?
And what I want you to do is to take maybe the next 5 or 10 minutes, make a list, and write down the materials and methods section for a paper on how to fold a paper airplane, okay? So, maybe give yourself five minutes and what I want you to do is go ahead and populate this file with instructions on how to fold a paper airplane.
Great. Hopefully, you now have instructions on how to fold a paper airplane. Now, if we go to the bottom of the page, you'll see a rectangle that says "Commit changes." In soft gray text, it says, "Update README.md."
So, I'm going to type something in here that's a little bit more informative than "Update README.md," but I'm going to say, "Write instructions to fold a paper airplane." That's it. I'm not going to put anything in this other box, and I'm going to hit "Commit changes," okay?
And so, now, we see the result is, "How to fold a paper airplane," and at least my instructions, okay? So, no cheating. Don't look at my instructions.
Exercise
- Send a link to your repository to a neighbor and ask them to follow the directions.
- Revise your instructions based on your neighbor's success or failure.
- "Commit" the change
Hopefully, you have your own instructions at this point, okay? So, what I'd like you to do then is take these instructions and…or take the link to these instructions from up here, say, and email that to a friend, and have them go through your instructions on how to fold a paper airplane.
Maybe have them, if they're in the lab with you, have them show you the plane, or if they're somewhere else, have them take a picture and send you the plane. And then ask yourself, does what they folded look like what you had in mind from your instructions, okay? And what you could then do is go ahead, come back to click "Edit this page," and in here, you could then modify your text to improve the reproducibility of your instructions.
You see what I did there? Hah. So, based on what they got right, or what they got wrong, or what they were confused about, modify your instructions to improve your instructions, okay? And I'm going to go ahead and say, "Obtain an 8.5" x 11" white piece of paper," and maybe down here I'll add 9.
I'll say, "Use crayons or sharpies to decorate your plane," okay? So, I've added some changes. I can then come back down to "Commit changes," and then I can say, "Added instructions to improve the decoration…" Or I'll say, "The design… The aesthetics of the paper airplane," okay?
So, they encourage me to use fewer than 50 characters. I'm not going to worry about that at this point. I'll go ahead and hit "Commit changes," okay? So, now I have an improved version of my instructions. And the cool thing about this is, as you saw by sharing this with your friend, anybody can see it. These instructions are publicly accessible to anybody.
The other thing we've seen is that we can constantly modify this. We can modify and improve our instructions over time. We could perhaps also click on this History button and see how our document has changed over time. And so, this, again, is very powerful in allowing us to disseminate, modify, improve our documentation.
So, I'm going to go head back to my rr_practice. The nice thing about GitHub is that if there's a file called README.md, it renders it on the homepage for that repository. Great. So, hopefully, this was useful in thinking about reproducibility.
You know, folding a paper airplane is something that at least a lot of Americans are familiar with. You might think about folding paper airplanes with people of other cultures and see if that's something they're used to. So, let's go ahead and come back to…let's come back to our slide deck and debrief a bit from that activity.
Debrief
- What were the challenges to reproducibly making a paper airplane?
Perhaps you found you had to be specific about the shape of the paper. Perhaps you had to think about, how do you describe the pointy end or the shape of the airplane at different stages? You might have a mental image of what that plane looks like whereas others don't have that same image.
Debrief
What were the challenges to reproducibly making a paper airplane?
What would improve your ability to improve the reproducibility?
Perhaps you found you had to be specific about the shape of the paper. Perhaps you had to think about, how do you describe the pointy end or the shape of the airplane at different stages? You might have a mental image of what that plane looks like whereas others don't have that same image.
I think a lot of us have folded paper airplanes in our childhood or playing with kids, and maybe others haven't, and so, if you've got that mental image, then you kind of assume things about people's prior knowledge. What could improve your ability to improve the reproducibility? Well, you could embed pictures in your README file, right? So, if you provided a more pictorial description of the method, then that would overcome a lot of problems. And so, think about, how does a tool like GitHub make your method more reproducible? Well, as we described, your friend can use it.
Debrief
What were the challenges to reproducibly making a paper airplane?
What would improve your ability to improve the reproducibility?
- How does a tool like GitHub help to make your method more reproducible?
Perhaps you found you had to be specific about the shape of the paper. Perhaps you had to think about, how do you describe the pointy end or the shape of the airplane at different stages? You might have a mental image of what that plane looks like whereas others don't have that same image.
I think a lot of us have folded paper airplanes in our childhood or playing with kids, and maybe others haven't, and so, if you've got that mental image, then you kind of assume things about people's prior knowledge. What could improve your ability to improve the reproducibility? Well, you could embed pictures in your README file, right? So, if you provided a more pictorial description of the method, then that would overcome a lot of problems. And so, think about, how does a tool like GitHub make your method more reproducible? Well, as we described, your friend can use it.
It's publicly accessible. Others could use it.
Debrief
What were the challenges to reproducibly making a paper airplane?
What would improve your ability to improve the reproducibility?
- How does a tool like GitHub help to make your method more reproducible?
- What might you do with your neighbor's repository?
Perhaps you found you had to be specific about the shape of the paper. Perhaps you had to think about, how do you describe the pointy end or the shape of the airplane at different stages? You might have a mental image of what that plane looks like whereas others don't have that same image.
I think a lot of us have folded paper airplanes in our childhood or playing with kids, and maybe others haven't, and so, if you've got that mental image, then you kind of assume things about people's prior knowledge. What could improve your ability to improve the reproducibility? Well, you could embed pictures in your README file, right? So, if you provided a more pictorial description of the method, then that would overcome a lot of problems. And so, think about, how does a tool like GitHub make your method more reproducible? Well, as we described, your friend can use it.
It's publicly accessible. Others could use it.
And something that you might also think about, well, say somebody sent you this repository. What would you do? What could you do with it, perhaps? Perhaps you could say, "Well, I'd like to get a bunch of different instructions for making different types of airplanes." I was always jealous by the kids that had really cool airplanes that were different than my kind of generic airplane, right? Perhaps you could compile a bunch of instructions to make an encyclopedia of paper airplanes, or perhaps you could take my boring paper airplane design and you could bling it up a bit. You could give it fins, or you could adjust the weighting, or talk about adding a paperclip to make sure the nose points down, or adjust the weighting, right? So, it's now open. It's something that you can now riff off of, as we've been saying in these tutorials.
Debrief
What were the challenges to reproducibly making a paper airplane?
What would improve your ability to improve the reproducibility?
- How does a tool like GitHub help to make your method more reproducible?
- What might you do with your neighbor's repository?
- Was this easy or difficult?
Perhaps you found you had to be specific about the shape of the paper. Perhaps you had to think about, how do you describe the pointy end or the shape of the airplane at different stages? You might have a mental image of what that plane looks like whereas others don't have that same image.
I think a lot of us have folded paper airplanes in our childhood or playing with kids, and maybe others haven't, and so, if you've got that mental image, then you kind of assume things about people's prior knowledge. What could improve your ability to improve the reproducibility? Well, you could embed pictures in your README file, right? So, if you provided a more pictorial description of the method, then that would overcome a lot of problems. And so, think about, how does a tool like GitHub make your method more reproducible? Well, as we described, your friend can use it.
It's publicly accessible. Others could use it.
And something that you might also think about, well, say somebody sent you this repository. What would you do? What could you do with it, perhaps? Perhaps you could say, "Well, I'd like to get a bunch of different instructions for making different types of airplanes." I was always jealous by the kids that had really cool airplanes that were different than my kind of generic airplane, right? Perhaps you could compile a bunch of instructions to make an encyclopedia of paper airplanes, or perhaps you could take my boring paper airplane design and you could bling it up a bit. You could give it fins, or you could adjust the weighting, or talk about adding a paperclip to make sure the nose points down, or adjust the weighting, right? So, it's now open. It's something that you can now riff off of, as we've been saying in these tutorials.
And so, was this easy or difficult? Right? I think you'll find that it's perhaps a little bit more difficult than you thought it was. You know, everyone knows what a paper airplane is, right? Well, everyone knows what PCR is, right?
Maybe you have certain assumptions about what other people know how to do, and those assumptions might not actually be correct. And so, where we're going again with this tutorial series is to making things as reproducible and automated as possible. And so, I found this video on YouTube that I think is pretty cool, that somebody programmatically engineered a way to fold a paper airplane, right?
So, you can imagine that people would…that if you want a reproducible airplane, this thing's going to give it to you, right? I'm sure they could churn out 100 paper airplanes in 1 minute, and they'd all be about the same, right? And so, that's really where we want to go with our data analysis.
Introspection
- Think about a figure from the last paper you wrote or that was in your favorite paper
- What would you need to tell someone else how to create the figure?
- What would be the benefits of creating a "script" to tell someone how to generate the figure?
- What would be the challenges?
So, what I'd like you to do, turning back to data analysis for microbiome data, is, think about a figure from the last paper you wrote or that was in your favorite paper. What would you need to tell someone else how to create that figure, right? So, instead of making instructions for a paper airplane, now what instructions would be needed to make that last figure?
What would be the benefits of creating a "script" to tell someone how to generate that figure, and what would be the challenges? Okay? So, again, these are topics that we're going to be going over in the rest of the tutorial series, but it's good to start to formulate ideas about your answers to these questions and how you would overcome them, and how not being able to answer them, or not being able to overcome them, perhaps limits our reproducibility, limits the ability of others to build off of our work.
Matching
Tool
- GitHub
- Markdown
- ORCID
- SRA
- FigShare
- MIMARKS
Application
- Minimal metadata standard
- Lightweight method to format text
- Database to post general data
- Sequence read archive
- Mechanism for linking papers to you
- Website to making code public
So, finally, I have a little exercise for you where I've got, on the left, six different tools, and on the right, six different applications, and I'm not going to do this as part of the video. The instructions are there if you hit the P key, or the answers are there if you hit the P key. But I'd like you to match the different tools with the different applications.
- GitHub - Website to making code public
- Markdown - Lightweight method to format text
- ORCID - Mechanism for linking papers to you
- SRA - Sequence read archive
- FigShare - Database to post general data
- MIMARKS - Minimal metadata standard
Well, I really hope that you enjoyed thinking more about some simple first steps that you can take to improve the reproducibility of your data analysis. As I mentioned in the last tutorial, if you make it all the way through the tutorial series, there will be an opportunity for you to receive a virtual badge indicating that you completed the material. To receive that badge, a later activity will ask you to demonstrate that you have done the activities in each of the tutorials, things like getting an ORCID and GitHub account, and posting the repository that contains the instructions for folding your paper airplane.
Today's motivation was thinking about incremental steps that you can take to improve the reproducibility of each analysis you work on. That being said, in the remaining sessions, the topics are going to get a bit more technical and a bit more involved, although, hopefully, they'll still be quite accessible, even to the novice. By the end of this tutorial series, and with much practice, you'll be what I call a full stack reproducible research data scientist.