class: middle center # Literate programming .footnote.left.title[http://www.riffomonas.org/reproducible_research/literate_programming/] .footnote.left.gray[Press 'h' to open the help menu for interacting with the slides] ??? Welcome back to the Riffomonas Reproducible Research Tutorial Series. I hope you're able to make it to the last tutorial where we added an R Script to our driver file to generate a figure with an ordination plot. I'm constantly amazed by the growing number of resources that are available in both R and Python to make our data analysis more reproducible. One of these new resources is R Markdown. This is a blend of Markdown, which we've already discussed when talking about documentation in R code which we discussed in the previous tutorial. R Markdown has revolutionized how my research group approaches scientific publisher. Emerging from the Python ecosystem, there's a similar tool called Jupyter Notebooks. Both R Markdown and Jupyter are examples of literate programming in which native text is blended with computer code, the results are pretty marvelous. Have you ever had to update numbers in a manuscript after changing a parameter, using a different statistical analysis, or adding more data? It's painful. More than one time, I've had errors slip into my manuscripts because I forgot to update all of the numbers. And tables with dozens of values, ah, what a pain. Literate programming removes that pain. If you look at my papers from my research group that are posted on GitHub, you can find R Markdown documents that contain R code behind any summary statistical P value. The tables are generated by R code. Furthermore, combined with the idea of a driver script, which we've been developing in this tutorial series, not only can you find the code for the summary statistic, but you can track it all the way back to a raw data file. If you remember back to the introduction for this series, I mentioned an April Fools' joke we played introducing a write.paper function into mother. Well, R Markdown is basically that function. I can't wait to show you how to write research reports and manuscripts using R Markdown. Join me now in opening the slides for today's tutorial which you can find within the Reproducible Research Tutorial Series on the riffomonas.org website. --- ## Pop quiz How do you run a function (e.g. `plot_nmds`) from R script (e.g. `plot_nmds.R`) at the bash shell prompt? <br> ??? Before we get going in discussing literate programming with R Markdown, I have a brief pop quiz for you that will hopefully jog your memory at how we can work with R from R pipeline. So, the question is, how would you run a function? So for example, we saw it before, plot_nmds from our R script which is also called plot_nmds.R at the bash shell prompt. Take a couple moments, and see if you can look back through your notes or if you remember off the top of your head how you would go ahead and run this function from this R script without first going into R. So, hopefully, that jogged your memory and you had some recollection of doing this in the previous tutorial. -- ```bash R -e "source('code/plot_nmds.R'); plot_nmds('data/mothur/stability.trim.contigs.good.unique.good.filter.unique.precluster.pick.pick.pick.opti_mcc.unique_list.thetayc.0.03.lt.ave.nmds.axes')" ``` ??? But recall that we can use R -e to execute a string of commands from the bash command line. And so that needs to be...those commands need to be embedded within single or double quotes, they need to match, and the individual commands can be separated with a semicolon. So here, bound within double quotes, we have two commands, we have a source function, and a plot_nmds function. The source function loads the code from code/plot_nmds.R. That file name is wrapped in single quotes. And then we run plot_nmds, and that takes a .axes file that is also wrapped in single quotes. Man, that's a long file name. So again, if we run this from the bash command prompt, we don't have to go into R, but the code will still get run and then it'll quit out of R. And so, this is really nice if we want to be able to automate our pipeline because we can run the R code without actually having to manually go into R. --- ## Learning goals * Express how a manuscript is an extension of programmatic analyis * Define literate programming * Demonstrate how to use code chunks and inline code to insert information into text * Implement advanced Rmarkdown features to add citations, figures, and tables * Manipulate YAML material to impact output format ??? For today's tutorial, I hope to help you learn how to express a manuscript as an extension of a programmatic analysis to really see that typing and writing our text, our narrative can be fused with programmatic analysis, and this is what we call literate programming. Within R Markdown, they use code chunks and inline code to insert information into the text. So, we'll talk about how to do that. And we'll also implement advanced R Markedown features including citations, figures, and tables to make it more of a polished manuscript. And then we'll talk about YAML and YAML material that can be used to impact the format of your output. --- class: middle center  ??? So, looking at this paragraph that you've already seen and copied into your main ReadMe file, this is the paragraph called Scaling up from the Kozich analysis. You'll notice that there's a variety of numbers in here that we had to calculate somewhere, right? So, down here, there's an error rate of 0.07% for the 2 mock communities, another 0.01%for the curation, and we had 14,094 sequences. All of these numbers would need to be updated if we change the data sets, if we changed how we calculated error, if we changed any steps in the pipeline, right? And if you look at this one paragraph, there's maybe a dozen or so different numbers that are being generated elsewhere, right? --- class: middle center  ??? And so, here in these red rectangles are those numbers that we had to calculate somewhere, right? So whether it's the number of samples, a citation number, numbers of sequences, numbers of reads per sample, and so forth, P-values that we had to generate somewhere and then insert into the text. --- class: middle center  .left.alert[ * Who wants to update these numbers if we change an upstream parameter? * What if we were to add a sequencing run? ] ??? Similarly, here's a table from that same paper where there's maybe 100 or so different numbers that if we, again, change the pipeline or if we added another run of data, we would have to update this table, and that could just be a royal pain, because you're sure to introduce all sorts of errors in doing that. --- ## To recap .left-column[ * Counts * Calculated values * P-values * References * Figure numbers * Figures * Tables ] .right-column.middle[ <br> .alert[Each of these can be addressed with ***Literate Programming***] ] ??? And so to think about what goes into a paper beyond the narrative, we have things like counts, the numbers of things, we have calculated values, we have P-values, we have references, figure numbers, the actual figures, and tables themselves. Each of these can be added and addressed into a paper, into a manuscript, using literate programming. --- ## Literate programming * Merging coding with text generation and formatting * Developed and championed by Donald Knuth * Several modern options * Jupyter Notebooks * RMarkdown notebooks and documents * Many types of output: markdown, PDF, docx, html ??? Literate programming is the idea that we can merge code with text generation and formatting. Literate programming was developed and championed by a world-renowned computer scientist named Donald Knuth. And currently, there are several modern options. So, as I mentioned in my introductory remarks, there is Jupyter and R Markdown from the Python and R environments, respectively. From these types of literate programming, we can get many types of output, whether it's a Markdown, plain text file, a PDF, a word docx file, or HTML code for rendering on a website. --- ## Applications of literate programming * Papers: [`rmarkdown`](http://rmarkdown.rstudio.com), [`knitr`](https://yihui.name/knitr/) * Books: [`bookdown`](https://bookdown.org) * Slide decks: [`slidify`](http://slidify.org), [`slidy`](http://rmarkdown.rstudio.com/slidy_presentation_format.html), [`ioslides`](http://rmarkdown.rstudio.com/ioslides_presentation_format.html) * Blogs: [`blogdown`](https://github.com/rstudio/blogdown) * Interactive websites: [`htmlwidgets`](http://www.htmlwidgets.org), [`shiny`](http://rmarkdown.rstudio.com/authoring_shiny.html) ??? Thinking about R Markdown, there's a lot of applications that have come out of the ability to use R Markdown in the packages R Markdown and Knitr, which are two packages we'll use that work together. People have used R Markdown to write books, create slide decks, blogs, and interactive websites. It's been really powerful. I liken it to a cookbook that if you go into Amazon and look at the reviews for a random cookbook, invariably, you'll find reviews that say this cookbook described how to make these types of cookies but the cookies taste awful. Well, think about a slide deck where your teaching R or you're showing somebody your code and your figure, it's like giving someone the cookie and the recipe used to make the cookie, right, well we can have the plot or the P-value and the code right there with it to know how those values are calculated or how those images were generated. I've taught entire semester-long courses using slide decks built using R Markdown, and why we're here discussing this today is that I write now manuscripts entirely in R Markdown. --- class: middle center  ??? And again, if we think about this paragraph and the various values that we might want to calculate using R --- class: middle ```R ***Scaling up.*** The advantage of the dual index approach is that a large number of samples can be sequenced using a number of primers equal to only twice the square root of the number of samples. To fully evaluate this approach, we re-sequenced the V4 region of 360 samples that were previously described by sequencing the distal end of the V35 region on the 454 GS-FLX Titanium platform ([18](#_ENREF_18)). In that study we observed a clear separation between murine fecal samples obtained from 8 C57BL/6 mice at 0 to 9 (“early”) and 141 to 150 (“late”) days after weaning and there was significantly less variation between the late samples compared to the early samples. In addition to the mouse fecal samples, we allocated 2 pairs of indices to re-sequence our mock community. We generated 4.3 million pairs of sequence reads from the 16S rRNA gene with an average coverage of `r round(mean(nseqs), 0)` pairs of reads per sample (95% of the samples had more than `r round(quantile(nseqs, 0.05), 0)` pairs of sequences) using a new collection of 8-nt indices (Supplemental Materials). Although individual samples were expected to have varying amplification efficiencies, analysis of the number of reads per index did not suggest a systematic positive or negative amplification biases that could be attributed to the indices. The combined error rate for the two mock communities was 0.07% before pre-clustering and `r format(pc_error_rate, nsmall=2)`% after (n=`r pc_error_nseqs` sequences). When we used UCHIME to remove chimeras and rarefied to 5,000 sequences there were an average of `r format(n_otus, nsmall=1)` OTUs (i.e. `r format(n_otus-perfect, nsmall=1)` spurious OTUs). Similar to our previous results, ordination of the mouse fecal samples again showed the separation between the early and late periods and increased stabilization with age (Figure 4; Mantel Test Coefficient= `r format(round(m.test$statistic, digits=2), nsmall=2)`, p\<`r format(round(m.test$signif, digits=3), nsmall=3)`**). These results clearly indicate that our approach can be scaled to multiplex large numbers of samples. ``` ??? We can now look at this example of R Markdown. Not all of the numbers have been converted to R Markdown. But you might look over here and you'll see these backticks with R and then R code embedded. This R code, when it's rendered by Knitr and R Markdown together, will spit out a number telling the reader how many pairs of reads there were per sample. And so, this is going to be pretty foreign to you right now but by the end of this tutorial, hopefully, you'll understand what's going on here. --- ## RMarkdown * Recall we previously used markdown in the paper airplane example and to write our README files * We have the ability to insert R code to generate tables, figures, and text * `rmarkdown` package uses the `knitr` package and other goodies to convert Rmarkdown files to a variety of formats ??? So hopefully, you recall that we previously used Markdown in the paper airplane example and in our ReadMe files for providing documentation. R Markdown is the idea that we can take that Markdown and embed R code into it to generate text, tables, and figures. The R Markdown package from R uses the Knitr package and other goodies to convert R Markdown files into a variety of formats. --- class: middle, center  .left.footnote[Source: [RStudio](http://rmarkdown.rstudio.com/lesson-2.html)] ??? So, you can see the schematic from RStudio. We write in R Markdown, used Knitr to convert that to Markdown and then a program called Pandoc will convert that Markdown into a variety of formats. This whole pipeline is fairly opaque to you. All you need to worry about is writing the R Markdown, setting what the output you want it to be, and you'll get it. And that is all done using the programs Knitr and R Markdown. --- ## Output for papers * Markdown - basic text * HTML - really nice for lab reports - limitless formatting (via HTML/CSS) * docx - good for manuscripts, easy for collaborators to work with - limited formatting (via `template.docx`) * PDF - good for manuscripts, harder for collaborators to work with - limitless formatting (via LaTeX; `new_project` helps) ??? So the outputs we can get include Markdown for basic text. And again, if we were to look at this on some site like GitHub where it automatically converts Markdown to HTML, that would serve our purposes. Alternatively, we can generate HTML-based websites. This is really nice for lab reports and affords you limitless formatting options using HTML, CSS, JavaScript if you want. You can also have things outputted as a docx file. This is great for manuscripts. We find that it's easier for our collaborators to work with the docx file than the R Markdown file. There is a bit of a limited formatting issue. You can format using a template.docx file, wherein that template file, you provide the formatting that you want. Things like, what font do you use, what size, double spacing, line numbering, things like that. We can also output PDFs. I find that this is my preference for manuscripts but I find that it's harder for my collaborators to work with because they tend to want to get into the text and mess with it and do things like track changes and all the things they're used to doing. But there is then limitless formatting via LaTeX. And so, you don't need to know LaTeX to generate a PDF, but it does then allow you to have more options in formatting. There are various helpers within the new_project directory template that we'll talk about later. And so, again, there's a range of outputs for papers. You need to find what works best for you and your collaborators in terms of the output. --- ## Note * You can do all of this through RStudio - they also have a notebook-like interface * We aren't going to do that because we are... * trying to emphasize the importance of having a workflow that is automated and can run without your intervention * assuming you will likely want to run everything on a cluster and may not be able to run a GUI on your server ??? So, we can do all of this through RStudio. They also have a nice notebook-like interface, and they have various helpers to make things easier, but we're not going to use RStudio. I'm sorry. Because we're trying to emphasize the importance of having a workflow that's automated and can run without our intervention, having to then go into a graphical interface kind of limits that. I'm also assuming that you'll likely want to run everything on a cluster and may not be able to run a graphical interface on your server. I know that there's a huge barrier to using a graphical user interface on my Local Flux cluster or even going on to Amazon. --- ## Returning to Kozich * Log in to AWS instance and start FileZilla * In the tutorial on organization you copied the "Scaling up" paragraph and the figure legend into your project's README.md file * Move this text into a new file `submission/practice.Rmd` ??? We're going to do is to return to our Kozich analysis. We're first going to log into our instance and start FileZilla so it'll be easier to transfer files back and forth and see what's being generated. Previously in the tutorial on organization, you copied the Scaling up paragraph into your README file. We're going to move that now to our submission/practice.Rmd file. --- class: middle ```bash **Scaling up.** The advantage of the dual-index approach is that a large number of samples can be sequenced using a number of primers equal to only twice the square root of the number of samples. To fully evaluate this approach, we resequenced the V4 region of 360 samples that were previously described by sequencing the distal end of the V35 region on the 454 GS-FLX Titanium platform (18). In that study, we observed a clear separation between murine fecal samples obtained from 8 C57BL/6 mice at 0 to 9 (early) and 141 to 150 (late) days after weaning, and there was significantly less variation between the late samples than the early samples. In addition to the mouse fecal samples, we allocated 2 pairs of indices to resequence our mock community. We generated 4.3 million pairs of sequence reads from the 16S rRNA gene with an average coverage of 9,913 pairs of reads per sample (95% of the samples had more than 2,454 pairs of sequences) using a new collection of 8-nt indices (see the supplemental material). Although individual samples were expected to have various amplification efficiencies, analysis of the number of reads per index did not suggest a systematic positive or negative amplification bias that could be attributed to the indices. The combined error rate for the two mock communities was 0.07% before preclustering and 0.01% after (n = 14,094 sequences). When we used UCHIME to remove chimeras and rarefied to 5,000 sequences, there was a average of 30.4 OTUs (i.e., 10.4 spurious OTUs). Similar to our previous results, ordination of the mouse fecal samples again showed the separation between the early and late periods and increased stabilization with age (Fig. 4) (Mantel test coefficient, 0.81; P < 0.001). These results clearly indicate that our approach can be scaled to multiplex large numbers of samples. **FIG 4** Principal coordinate ordination of YC values (28) relating the community structures of the fecal microbiota from 12 mice collected on days 0 through 9 (Early) and days 141 through 150 (Late) after weaning. ``` .footnote[`submission/practice.Rmd`] ??? So this practice.Rmd file's going to be a new file that we'll generate and we'll use for practicing various aspects of using R Markdown. So, I've been able to log into my instance, so I'm going to go ahead and move to my Kozich_Re-analysis_AEM_2013 directory. So again, I'm going to open my README file. And I see my Scaling up paragraph here. And I'm going to copy that, CTRL+X out, and I said, nano submission/practice.Rmd. And that gets me in my Scaling up. And I forget if my README also had the figure legend. Doesn't look like it. So, I'm going to open back up submission/practice.Rmd, and I'm going to copy the figure legend for figure four. So again, this is a bit artificial because rarely would we want to go back and add the R code. You certainly can but it's much easier to write the R code as you're actually writing the paper. --- ## Getting set From within R ```R install.packages(c('rmarkdown', 'knitr')) # or # update.packages(c('rmarkdown', 'knitr')) ``` --- The `.Rprofile` file with `new_project` ```R ############################################################ ## Options for knitr and Rmarkdown rendering ############################################################ library("knitr") library("rmarkdown") ## output directory for figures if (require("knitr")) { opts_chunk$set(fig.path="results/figures/") opts_knit$set(base.dir=normalizePath(getwd())) opts_knit$set(root.dir=normalizePath(getwd())) } ############################################################ ## the following is credit to Jenny Bryan, for details see here: ## https://gist.github.com/jennybc/362f52446fe1ebc4c49f RPROJ <- list(PROJHOME = normalizePath(getwd())) attach(RPROJ) cat('Project home directory is available as PROJHOME or via get("PROJHOME","RPROJ")\n') rm(RPROJ) ############################################################ local({ r <- getOption("repos") r["CRAN"] <- "http://cran.cnr.berkeley.edu/" options(repos = r) }) ``` This will automatically load the libraries whenever you run R ??? Okay. I'll save this and exit out. Next, we'd like to go ahead and look at our .Rprofile file. So, if we do ls, you'll notice we don't see a .Rprofile file, but if you do ls -a, in here, the bottom right, we have a .Rprofile file. This is the file that is loaded whenever we run R. Now, we talked about in the last tutorial that we don't want to put a lot of stuff into this .Rprofile file. So let's see what's in there. So, did nano .Rprofile, and it says library Knitr, library R Markdown, and then it sets paths for where to put things from Knitr and from R Markdown, and it helps to normalize our path and to get things in the right place. So everything looks good here. One other thing I'll add is that the AWS instance you have has Knitr and R Markdown already installed, so you don't need to install packages for R Markdown or Knitr. We'll go ahead and reopen our submission/practice.Rmd file. And we're gonna start the process of converting this into an R Markdown document. --- ## Converting `md` to `Rmd` * At a minimum need to add a [YAML](http://www.yaml.org) header * Do not start lines with tabs, use spaces * Set apart from rest of document with opening `---` and closing `---` on their own lines <br> ```bash --- output: html_document --- **Scaling up.** The advantage of the dual-index approach is that a large number of samples can be sequenced using a number of primers equal to only twice the square root of the number of samples. To fully evaluate this <text continues...> ``` ??? At a minimum, to make it an R Markdown document, we need to add what's called a YAML header. So a YAML header is denoted by having three hyphens on the first line, some information, and then three hyphens to close the YAML header. In between the three hyphens, we need to add information that tells R Markdown and Knitr and Pandoc, eventually, what exactly needs to be done. So in here, we'll put output: html_document. So our YAML headers will get a little bit more complicated as we go along, but it's important to note that we never want to put a tab in our YAML header. --- Generate the HTML output like so... ```bash $ R -e "render('submission/practice.Rmd')" R version 3.3.2 (2016-10-31) -- "Sincere Pumpkin Patch" Copyright (C) 2016 The R Foundation for Statistical Computing Platform: x86_64-apple-darwin13.4.0 (64-bit) R is free software and comes with ABSOLUTELY NO WARRANTY. You are welcome to redistribute it under certain conditions. Type 'license()' or 'licence()' for distribution details. Natural language support but running in an English locale R is a collaborative project with many contributors. Type 'contributors()' for more information and 'citation()' on how to cite R or R packages in publications. Type 'demo()' for some demos, 'help()' for on-line help, or 'help.start()' for an HTML browser interface to help. Type 'q()' to quit R. Project home directory is available as PROJHOME or via get("PROJHOME","RPROJ") > render('submission/practice.Rmd') processing file: practice.Rmd |.................................................................| 100% ordinary text without R code output file: practice.knit.md /usr/local/bin/pandoc +RTS -K512m -RTS practice.utf8.md --to html --from markdown+autolink_bare_uris+ascii_identifiers+tex_math_single_backslash --output practice.html --smart --email-obfuscation none --self-contained --standalone --section-divs --template /Library/Frameworks/R.framework/Versions/3.3/Resources/library/rmarkdown/rmd/h/default.html --no-highlight --variable highlightjs=1 --variable 'theme:bootstrap' --include-in-header /var/folders/8p/j4r5_4yn7jg5z3s6p_ky8lc40000gq/T//RtmpsTef9B/rmarkdown-str14c9de98510.html --mathjax --variable 'mathjax-url:https://cdn.mathjax.org/mathjax/latest/MathJax.js?config=TeX-AMS-MML_HTMLorMML' Output created: practice.html ``` ??? Any kind of justification along the left side, left margin, we want to do with spaces. So, we'll go ahead and save and quit this. And so then, from our command line, we can render this doing R -e "render('submission/practice.Rmd')". Remember, we don't need to put library R Markdown or library Knitr in here because that's automatically loaded as part of our Rprofile file. So we'll run this. And it runs pretty quickly. --- class: middle center  .left.footnote[`submission/practice.html`] ??? And we see, "Output created: practice.html." So I need to connect to my AWS instance in FileZilla. And I need to update my IP address and I'm going to get that from my AWS instance. Copy it to the clipboard, come back over here, paste that in, connect. Yes, I'll trust them, OK. Kozich_re-analysis, submission, practiced.html, opening that up. I see I now have a HTML formatted version of the Scaling up file. That's great. So at a basic level we see that it works. --- ## Let's add a title, date, and author </br> ```bash --- title: Reproducing Kozich et al author: Joe Schmo date: "February 30, 2017" output: html_document --- **Scaling up.** The advantage of the dual-index approach is that a large number of samples can be sequenced using a number of primers equal to only twice the square root of the number of samples. To fully evaluate this <text continues...> ``` ??? So let's add a title, date, and author to our YAML. So, I'm going to reopen my submission/practice file, and I'm going to add title:, and I'll say, Reproducing Kozich et al. Author, I'm going to put my name in, you put your name in, Pat Schloss. Date, and I'm going to say April 24th, 2018. And I'll go ahead and save this and quit. And then back out in bash, I'm going to render this, and I can then go to FileZilla, I can refresh, and refresh here. Maybe I need to double-click again on it. Reopen local file. Discard local file, then download and edit file anew. Okay. --- class: middle, center  ??? So now, we see we have a title. Lost the al. My name and my date. All right. So it nicely puts in this information that we added into the YAML header. --- Can we automate these calculations? ```bash mock community. We generated 4.3 million pairs of sequence reads from the 16S rRNA gene with an average coverage of 9,913 pairs of reads per sample (95% of the samples had more than 2,454 pairs of sequences) using a new ``` ??? So, I'll return to my Rmd file. And if we look down in here, we see that there's a couple of numbers here, the 4.3 million pairs of sequence reads from the 16S RNA gene with an average coverage of 9,913 pairs of reads per sample. With...95% percent of the samples had more than 2,454 pairs of sequences. Okay. What I'd like to do is to automate those calculations in my Rmd file. So, to do this, we'll need to create what's called a code chunk. And we'll also need to make use of what's called inline code. -- <br> To do this, we'll need to create a ***code chunk*** ``` ```{r} # your code goes here `````` ??? A code chunk is denoted in R Markdown by ```{r}. And then the code chunk ends with another set of three backticks. So if you have a bunch of paragraphs in your paper, you can have code chunks scattered throughout your document. --- <br> ... and make use of ***inline R code*** ```R ...coverage of `r ave_number_of_pairs` pairs of reads... ``` ??? Because our document's pretty small here, we're only going to have one code chunk. --- Code chunk... ``` --- title: Reproducing Kozich et al author: Joe Schmo date: "February 30, 2017" output: html_document --- ```{r} shared_file_name <- "data/mothur/stability.trim.contigs.good.unique.good.filter.unique.precluster.pick.pick.pick.opti_mcc.unique_list.shared" shared_file <- read.table(file=shared_file_name, header=T, stringsAsFactors=F) sequence_counts <- rowSums(shared_file[,-c(1,2,3)]) million_sequence_counts <- sum(sequence_counts)/1e6 average_sequence_counts <- mean(sequence_counts) percentile_sequence_counts <- quantile(sequence_counts, prob=0.05) `````` ??? And so because this isn't an R course, I'm going to copy the notes...the code, I'm sorry, from the slide deck into the code chunk here. And so, we see...going to add some space in here so we can kind of differentiate what's going on. Okay. So we've got our shared filename, we're reading in our shared file with the R code, we're counting the number of sequences in each row, and then we're getting the sequence counts in millions by counting up the number of sequences divided by a million, the average sequence counts using the mean function, and then the percentile sequence counts here in this final line of the code chunk. Okay? So, this is the code that's going to be used later in the paragraph to embed into our sentence. And so this 4.3 million is going to come from this million sequence counts. -- Inline code... ```bash <snip> mock community. We generated `r million_sequence_counts` million pairs of sequence reads from the 16S rRNA gene with an average coverage of `r average_sequence_counts` pairs of reads per sample (95% of the samples had more than `r percentile_sequence_counts` pairs of sequences) using a new <snip> ``` ??? Okay. And so, these three variables hold information that's going to go into this paragraph using what I'm calling inline code. All right. So, 4.3 million pairs, I'm going to remove that 4.3 and put in a `R and I have another `. And so this means, between the backticks, we're gonna put R code that's going to be run and inserted directly into the sentence. So, I'm going to say, million_sequence_counts. All right. Similarly, down here we have average coverage. I'm going to replace this with `R, and inside the backticks, I'm going to put average_sequence_counts. And then in here, the 95% of samples have more than 2,454 pairs of sequences, I'm going to do, again, `r`, and inside the backticks, I'm going to put percentile_sequence_counts. All right. --- class: middle center  ??? So, see how we did that? We have a code chunk up here that reads in the file, it does the calculations. And then down here in the narrative part of the text, we can column these variables that were defined up here in the code chunk to be inserted indirectly. Now, we could have put everything in this code chunk directly into the inline code, but that would get really painful to read and difficult to edit later. And so again, we can use this code chunk to define variables that we then insert into the text. So, I'm going to go ahead and save this. And re-render. 'average_sequence_counts' not found. So, I'm going to check that out again. And I misspelled sequence up here in my code chunk. Save that, re-render. Everything worked. I'll come over to FileZilla, refresh, re-open, I'm going to discard, and then download the file anew. Aha. And so now what we see is that we have our code chunk, and then in this sentence where we put our R code, we see we generated 3.86 million pairs of sequence reads with an average coverage of 1.07 blah, blah, blah, times 10 to the 4 pairs of reads per sample. Okay. So the information is here but the formatting leaves a little bit to be desired. --- # .alert[ How well did we reproduce the previous results? ] ??? How well did we reproduce the previous results? Well, the numbers are pretty close. -- <br> # .alert[ Why would they be different? ] ??? The Kozich paper originally was not generated using an R Markdown document. They're pretty close. There were two data sets available, so it's possible we grabbed the wrong data set or that I used a different...there was one data set that was the with metagenomics and one was without the metagenomics. We may have grabbed the wrong one that I used originally. We could go back and check that out. There have also been software changes to mother over the years, that might cause some things. There's also some randomness at different steps in the pipeline. And it's also possible that the total number of sequences was really the number of raw sequences, and what we have here is the number of curated sequences that made it through our pipeline. And so these are all things that would have been nice if we would have documented it the first time around. We have since gone back and re-generated this paper using reproducible practices. --- ## Cool, eh!? * There are still a bunch of things we'd like to do with this before we submit it to a journal or even share with a collaborator. Let's think about what we've just done first * Code chunk - For a manuscript submission, we probably don't want to see the code - For our PI or select collaborators, we may want to show the code so they can proof our methods * Inline code - The number of significant digits is a bit much ??? And the numbers are pretty close and the gist of the story doesn't change. So this is really cool, right? There's still a bunch of things we'd like to do with us before we'd submit to a journal or even share it with a collaborator. And so, let's just think about what we've done first. Okay? We've created a code chunk and that reads in the file and it does some processing of the data. When we submit it, we probably don't want to see the code, we'd like to hide that. But for a PI or selected collaborators, we might want to show them the code so that we have proof of our methods and they can see what we've done. The inline code, the number of significant digits is a bit over the top. --- ## Code chunk options ``` ```{r scaling_up, echo=FALSE} shared_file_name <- "data/mothur/stability.trim.contigs.good.unique.good.filter.unique.precluster.pick.pick.pick.opti_mcc.unique_list.shared" shared_file <- read.table(file=shared_file_name, header=T, stringsAsFactors=F) sequence_counts <- rowSums(shared_file[,-c(1,2,3)]) million_sequence_counts <- sum(sequence_counts)/1e6 average_sequence_counts <- mean(sequence_counts) percentile_sequence_counts <- quantile(sequence_counts, prob=0.05) `````` ??? Okay? So, what we're going to first do is look at how we can get rid of seeing this code chunk. We still want it to run but we don't want to see it. So if we go back into our R Markdown document, inside our curly braces after the R, we can write echo=FALSE. What echo=FALSE will do, well, it will not echo the code. So, echo=TRUE is what we currently have where it echoes back the code to the screen. So we quit out, we re-render. -- This code chunk has done two things: * Names the code chunk (`scaling_up`), this helps with organizing our code chunks and figuring out where Bad Things Happen if `render` chrashes * `echo=FALSE` tells `knitr` to not output the code --- class: middle center  ??? And now, the code chunk is gone but the results of our R are still in the sentence. --- ## Other code chunk options that are useful * `include`: Tells whether code and results should be in the output (still runs code) * `echo`: Tells whether code, but not results should be in the output (still runs code) * `message`: Tells whether messages that are generated by code should be in the output * `warning`: Tells whether warnings that are generated by code should be in the output * [Many other options available](http://rmarkdown.rstudio.com/lesson-15.html) including how to format figures, caching, dependencies, etc. ??? So there are some other code chunk options that are useful. So include tells whether the code and results should be in the output. It will still run the code. Echo tells whether the code but not the results should be in the output. It will still run the code. Message, true or false, tells whether messages that are generated by the code should be in the output. And then, warning, true or false tells whether warning messages that are generated by the code should be in the output. In the slide deck, there's a link to many other options that are available for how to format figures, do things like caching, make dependencies between code chunks. But again, for most of what I do in my papers, I'm mainly using echo, message, and warning in my code chunks. --- ## Exercises * Create a new code chunk and in it add the line `summary(sequence_counts)`. What happens when you use `include` or `echo` and set the option to `TRUE` or `FALSE` * Create a new code chunk and insert a line with `hist(sequence_counts)`. Again, play with the `include` and `echo` options. Also adjust the `fig.caption`, `fig.height`, `fig.width`, `fig.align`, and `dev` parameters. Where does it put the figure? --- ## Formatting inline text ```bash <snip> mock community. We generated `r format(round(million_sequence_counts, 1), nsmall=1L)` million pairs of sequence reads from the 16S rRNA gene with an average coverage of `r format(round(average_sequence_counts, 1), nsmall=1L)` pairs of reads per sample (95% of the samples had more than `r format(round(percentile_sequence_counts, 1), nsmall=1L)` pairs of sequences) using a new <snip> ``` ??? One of the things that we tend not to think about when we use R a lot is how to format text or to format numbers. Normally, we're happy to pull a P-value, pull a summary statistics out of R, but with R Markdown, we really want to pull it into another document. So we need to think about how we format things. So returning to submission/practice. If I scroll down to where I embedded my R code, I probably want to round this number that we had as output. And we can do this with a function called round and format. And so I can say, format(round(million_sequence_counts. And so for rounding, I'm going to say, round to one significant digit. And then for format, I'm going to say, nsmall=1L). And what this will do is this will return a single digit...or a number with one significant digit nothing to the right of the decimal point. So if we save, render, and then open that up. So we see that we've got one significant digit to the right of the decimal point, 3.9, which is what we were hoping for, so that's a lot tidier. We can do the same type of thing...we can do the same type of thing with the other numbers that we generated. So if we go back to nano and we look at our average sequence counts, we can do the same thing where we round it to one significant digit format(round(average_sequence_counts, 1), nsmall=1L)`. And we'll do the same thing here with our percentile. We will do format(round. 1) 1L). Quit. -- <br> .center[] ??? Render. And so, we see a much nicer formatting. So we have 10,735.1 pairs as the average instead of, you know, something in scientific notation. And then 2,788.9 pairs of sequences. So in this sentence, we had 3 numbers, 3.9, 10,735.1, 2,788.9 that were all generated from R. If we return to our R code, and we looked down at this, we might notice that this isn't very dry, that we have the same format round and then the number and the same parameters over and over again. --- ## Put this after the YAML and before your first code chunk, what is it doing? ``` ```{r knitr_settings, eval=TRUE, echo=FALSE, cache=FALSE} opts_chunk$set("tidy" = TRUE) opts_chunk$set("echo" = FALSE) opts_chunk$set("eval" = TRUE) opts_chunk$set("warning" = FALSE) opts_chunk$set("cache" = FALSE) inline_hook <- function(x){ if(is.list(x)){ x <- unlist(x) } if(is.numeric(x)){ if(abs(x - round(x)) < .Machine$double.eps^0.5){ paste(format(x,big.mark=',', digits=0, scientific=FALSE)) } else { paste(format(x,big.mark=',', digits=1, nsmall=1, scientific=FALSE)) } } else { paste(x) } } knitr::knit_hooks$set(inline=inline_hook) `````` ??? And so, what we'd like to do is perhaps remove this so we don't have to repeat it but that our text can be formatted, our numbers can be formatted the same way every time we're outputting text. What we'll do is we'll go up and we'll create another code chunk. And I'm going to...we can name or code chunks, so I'm going to name this one Knitr settings. And I'm gonna say, eval=TRUE, so we're going to evaluate what's in here. Our echo is going to be false, and we're going to say, cache=FALSE. And we close that with three backticks. And in here I'm going to set several chunk options. So we can globally set our chunk options. So, I'm going to say, opts_chunk$set("tidy"-TRUE), and I'm going to copy this several times so I don't have to keep typing. So I'll do tidy=TRUE. Tidy refers to how the code is formatted. Echo=FALSE. We've already seen false echo where it outputs the code chunk. Eval=TRUE. Warning=FALSE. And cache=FALSE. You might want to run this a couple times with warning=TRUE. You might want to run once with warning=TRUE just to make sure you're not getting any problematic warning or error messages when you run your analysis. So if you look at the slide deck, there is a function in the slide deck called inline hook that I'm going to copy and paste into here. And this is a function that is run every time it does an inline code chunk. Okay? Wherever you see that `r and then R code, it runs this function on the contents within those backticks. And so, you'll see that there is information in here on what to do. Some of this we don't need to worry about. Is it a list, unlist it and spit it out as a vector, but if it's numeric, then it does different things. So it will format it with commas and no special digits for formatting. If it's not scientific or if it's not an integer, it will, again, use commas and it will use one significant digit and it won't use scientific notation. So this first one is saying is it an integer? If it's an integer, don't use a decimal. If it's not an integer, then use one significant digit. And so what this allows us to do then is to go back into our inline code, and to reformat the text. The other thing is that this code chunk would be a great place to load any libraries that we had to run. So we could source any utilities or we could library any packages that we're using in here. -- This would be a good place to put `library(xxxxx)` calls --- ## Re-Formatting inline text ```bash <snip> mock community. We generated `r million_sequence_counts` million pairs of sequence reads from the 16S rRNA gene with an average coverage of `r average_sequence_counts` pairs of reads per sample (95% of the samples had more than `r percentile_sequence_counts` pairs of sequences) using a new <snip> ``` ??? And so again, we can come down to our text, and we can remove these format round function calls. And from here. And here. -- <br> .center[] ??? So, we can save this and exit out, render it. And we see that we have the same formatting of our text but the text is much more dry. --- ## Other things we want to deal with... * Figures * Tables * References * Other output formats --- ## Figures * For a manuscript, it is preferred to generate the figures outside of the Rmd file * At some point, you'll have to upload your figures separtely from the manuscript * Markdown gives you the ability to insert figures with<br>`` ??? Some other things we'll want to deal with include figures and tables, references, and then other output formats. --- ## Figures ```bash indicate that our approach can be scaled to multiplex large numbers of samples.  **FIG 4** Principal coordinate ordination of YC values (28) relating the community structures of the fecal microbiota from 12 mice collected on days 0 through 9 (Early) and days 141 through 150 (Late) after weaning. ``` ??? And so, that's what we're going to spend the rest of this tutorial discussing. My preference in working with figures in a manuscript is to not generate the figure within my Rmd file. Well, if you're using an Rmd file as a notebook, like we saw in the Meadow et al paper, then that makes sense to leave it in there. But normally, when I'm submitting a manuscript, I have to submit the figure separate from the text. And also, sometimes the figures require a fair amount of code to generate, and I like to encapsulate that away into a separate file. But we can still see the figures show up in our output from our Rmd file. And that is using R Markdown code. So let's go back into our practice Rmd file. And I'm going to come to the bottom where I have my figure legends, and under that we can use markdown to insert an image. So I'm going to...and the syntax is...we type and then I'll talk. So the syntax is, as you see here, in that we use an exclamation point, square brackets, and inside the square bracket is the description. If you're familiar with HTML, this is like the alt attribute for an image tag, and then we give the path to get to the figure. All right. And so, I'm going to change my description to be figure 1. And then the path to figure is going to be a little bit weird because we need to give it a relative path to our submission directory. So we're going to do ../results/figures/nmds_figure.png. And if we save this and render, hopefully, that figure will now be inserted into the output of our Rmd file. -- .center[] ??? And sure enough, there is our figure. Now I could get rid of the text inside the description and that figure 1 will go away, because I don't really want to see that. And again, it's gone now. Okay? So that's how I tend to insert figures if I'm gonna put figures in the manuscript. Again, when you submit a manuscript frequently, you're not going to put the figures in with the figure legends, but if you're doing it for a report to give to your PI or a draft, then it's nice to be able to put the figures in with the manuscript. --- ## Tables * Some of the examples that were shown earlier were tables and they helped to motivate the need for literate programming * There are several packages for generating tables in R * [`xtable`](https://cran.r-project.org/web/packages/xtable/): best for compmlex tables; produces LaTeX that can be converted to PDF or HTML; can induce headaches * [`knitr`](): `kable` function produces nice simple tables; will work well with word, pdf, html --- ## Silly example... ``` ```{r, result="asis"} days <- as.numeric(gsub(".*D(\\d*)", "\\1", shared_file$Group)) n_samples_per_day <- numeric() mean_seqs_per_day_per_sample <- numeric() total_seqs_per_day <- numeric() unique_days <- sort(unique(days)) for(d in unique_days){ n_samples_per_day[as.character(d)] <- nrow(shared_file[days == d, -c(1,2,3)]) total_seqs_per_day[as.character(d)] <- sum(shared_file[days == d, -c(1,2,3)]) mean_seqs_per_day_per_sample[as.character(d)] <- mean(apply(shared_file[days == d, -c(1,2,3)], 1, sum), na.rm=T) } data <- data.frame(unique_days, mean_seqs_per_day_per_sample, total_seqs_per_day, n_samples_per_day) kable(data, digits=c(0, 1, 0, 0), row.names=FALSE, col.names=c("Day", "Seqs. per<br>sample (mean)", "Seqs. per<br>day (total)", "Number of<br>samples"), align='c') `````` ??? So we're going to go ahead and add a silly table to our Markdown document. And I'm going to put this at the end because normally, I have tables at the end of my manuscripts and because this isn't a tutorial on R necessarily, I'm going to go ahead and create a code chunk and copy the code over from the slide deck. So there's my code chunk frame and I'll add this as a table 1, is the name of my code chunk. The names aren't critical. Sometimes, it's helpful for debugging where things go wrong because it'll give you an error message of where problems happened. So I'm going to go ahead and copy and paste this code from the slide decks over in, and you can see what's...perhaps, what's happening is that we make a column for days, number of samples for days, mean samples per day per sample, and the total sequences per days, and then we have a loop that goes through and counts these things. But then...we get to the table is that it makes a data frame that has four columns. Unique days, so what are the days of the study. The mean sequences per day per sample, so how many sequences we have per day per sample. The total sequences by day, and then the number of samples we have by day. This then gets us into cable which is that package...that function, I'm sorry, in R Markdown that will build a nice table for us. We give it our name of our data frame. Next comes digits, which is the numbers to the right of the decimal point that we're going to use in formatting our table. We're not going to use any row names. Our column names are gonna be Day, Sqs. per sample (mean). BR, there's a bit of HTML to put in a line break, Sqs. per day (total), and Number of samples. Finally, we tell cable to align our columns to be the center. And so just to show you a little, we can give this as a string where each letter in the string tells you whether...tells Knitr whether it should be center, right, left, center, whatever, where you want the number positioned horizontally within the table. --- .center[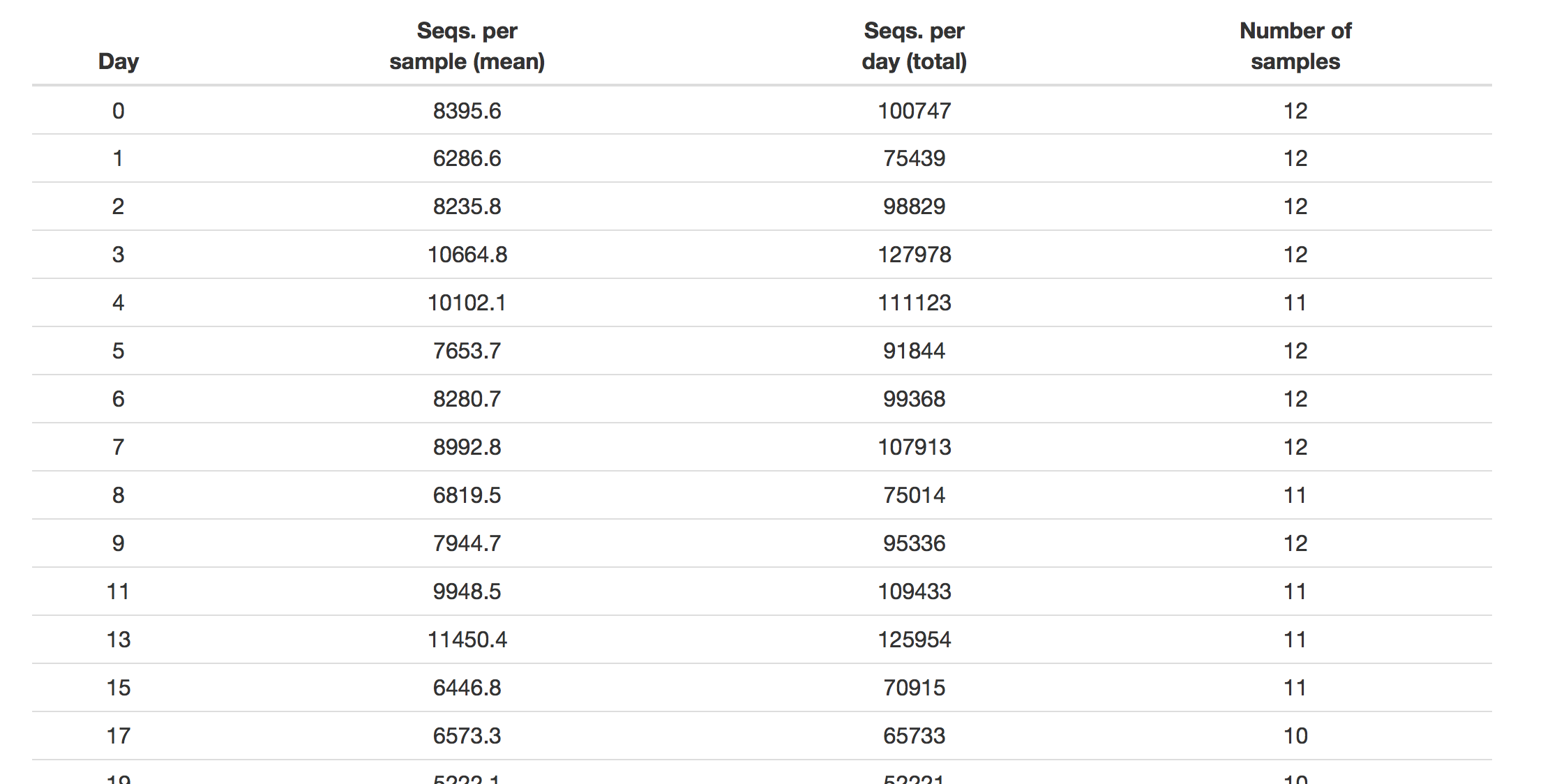] <br> More sophisticated tables can be generated using `xtable`. Consult the [xtable gallery](https://cran.r-project.org/web/packages/xtable/vignettes/xtableGallery.pdf) for examples and code ??? So if we save this, we render it. We then load it from FileZilla. And we see that we get a data frame. So, if you'll recall, we've made the first column left formatted, center, right, center. Okay. And also, we said zero digits to the right for the day 1, 1 to the right for column 2, 0 and 0 for the other 2 columns as well. So we can put in some nice formatting. And again, this is all HTML. And if you know some CSS, you could format this to look a bit more like a bit more polished, a bit more like you would like it to look. But again, this is a bit of a silly example telling us for each day of the mouse study, how many sequences we have per sample, the total number of sequences across all of our samples for that day, and the number of samples that we have. So you'll see that, you know, on day 302, we had 1 sample, but on day 0, we had 12 samples from our 12 different mice. --- ## References * RMarkdown allows you to use various citation managers to render references in your documents * Various formatting files are available as `csl` files that will play nicely with rmakrdown, Word, and LaTeX. You can find these files at the [CSL GitHub repository](https://github.com/citation-style-language/styles). As an example, we commonly use style guides for the [ASM Journals](https://github.com/citation-style-language/styles/search?q=asm&type=Code&utf8=✓) * A copy of `mbio.csl` is in `submission/` ??? So again, this is how we can put a figure and now a table into our R Markdown document. The next thing that we want to be able to do is to put references inside of our R code. And one of the things that we need to tell it is what type of formatting we want to use. And so, there's a variety that will work with R Markdown, Word, and LaTeX. And so, we can find these in what's called the CSL GitHub Repository, which you can find in the slide deck a link to. We commonly use style guides for the ASM Journals, and there's a copy of that called mbio.csl in the directory of CSL. So, if we...submission...you'll see here there's mbio.csl. And so, we can see the CSL GitHub Repository. We can see the CSL Repository by going to the GitHub Repository citation-style-languages/styles, and you'll see in here any number of journals represented, and you can search for the one you want because there's so many of them up here. So, if we did... I spelled American wrong. American...nope, that's right. And so, we see one for applied environmental microbiology or a variety of the ASM Journals in here. So if we wanted applied environmental microbiology, we would copy this file, and then pop this into our submission directory and we would name it. And so, this is what we've done but with the mbio.csl. And returning to our R Markdown document, we'll need to make a couple of changes. --- ## Need to make a couple of changes 1\. YAML ```bash --- title: Reproducing Kozich et al author: Joe Schmo date: "February 30, 2017" output: html_document bibliography: references.bib csl: mbio.csl #Get themes at https://github.com/citation-style-language/styles --- ``` ??? To our...to add to our YAML header, we'll say, csl: mbio.csl. And then we also need to tell it where our bibliography is. So I'll do bibliography: references.bib. And we'll save that. --- 2\. Create `submission/references.bib` ```bash @article{Schloss2012, doi = {10.4161/gmic.21008}, url = {https://doi.org/10.4161%2Fgmic.21008}, year = {2012}, month = {jul}, publisher = {Informa {UK} Limited}, volume = {3}, number = {4}, pages = {383--393}, author = {Patrick D. Schloss and Alyxandria M. Schubert and Joseph P. Zackular and Kathryn D. Iverson and Vincent B. Young and Joseph F. Petrosino}, title = {Stabilization of the murine gut microbiome following weaning}, journal = {Gut Microbes} } @article{Yue2005, title={A similarity measure based on species proportions}, author={Yue, Jack C and Clayton, Murray K}, journal={Communications in Statistics-Theory and Methods}, volume={34}, number={11}, pages={2123--2131}, year={2005}, publisher={Taylor \& Francis} } ``` * These are `bibtex`-formatted (*.bib) references. Can obtain from online tools such as [doi2bib](http://www.doi2bib.org/#/doi) or [google scholar](https://scholar.google.com) * Will use the `Schloss2012` as the tag that we cite ??? But now, we need to create references.bib and we'll also have to insert our references into our text here. So we'll quit out of that, and we will create a reference file. We'll do submission/references. Sorry, nano submission/references.bib. And by default, we put in here the mother paper, so, Schloss2009. And this is the type of formatting that we need to represent our papers. And so, I want to get two out of here. And so, one is the original paper that these data were taken from. And so I will go to PubMed and the authors on that were Schloss, Schubert, Zackular. And that's this. And I'm going to copy this DOI number, and there's a nice tool called DOI to bib.org and can give it a DOI, a PMCID, or an archive ID. And so if I hit enter on that, I get nice, BibTeX formatted material. So I can copy that and I can go back in here and I can paste that in. And so now, I've got these two and the other file that I want is...the other paper, sorry, that I want is...if I go back to PubMed, is Clayton and MK, Yue, JC. And this was the [inaudible]. And so we got to get the DOI number. So, I'll click on the paper. There's the DOI. And so I can click this DOI and go back to doi2bib. And there we go. So, I can copy that now into my references.bib file. Okay. --- 3\. Insert reference into Rmd file ```bash <snip> Titanium platform [@Schloss2012]. In that study, we observed a clear separation <snip> **FIG 4** Principal coordinate ordination of YC values [@Yue2005] relating the <snip> ``` ??? And now I have three files or three papers represented in my bib file. So to cite these in our manuscript, we now want to go back into submission/practice.Rmd and we want to replace this 18 with our reference. So we'll do [@Schloss2012]. And this is coming from our references file. So if you scroll down, till we had Schloss2012, this is the name that we're going to cite,Schloss2012. Similarly, if we wanted to use...cite the [inaudible], this Yue2001 would be what we cite. So let's go ahead and put that in. And scrolling to the bottom and we have this 28. We're going to see that that was @Yue2001. -- <br> 4\. Render .center[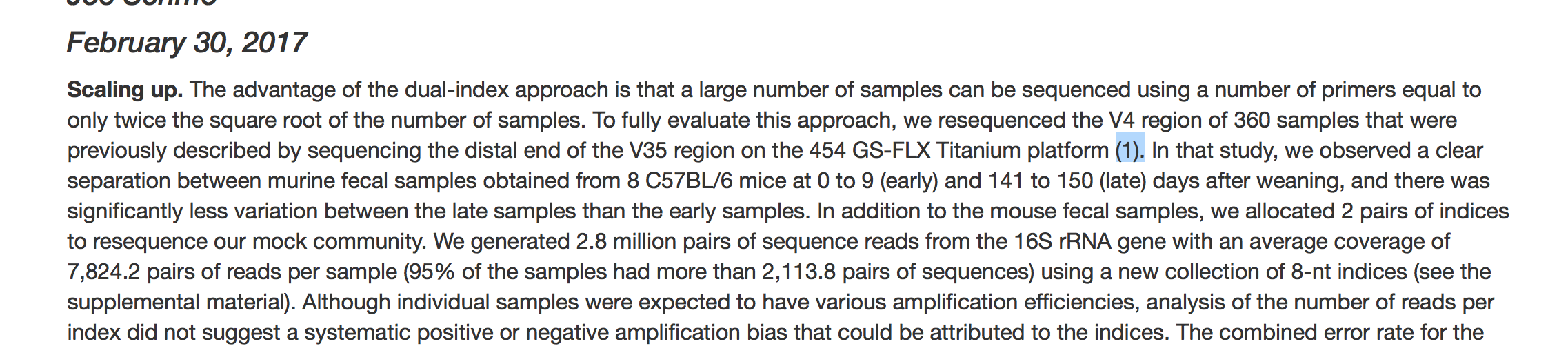] -- .center[] ??? I'll quit that and we can then render it. This runs through. We then open up this and we see Yue Clayton has a (2) and the original mouse study has a (1). And if we scroll to the bottom, we see our references. So maybe we'd want to say...if you'd want to add, say nano submission at the bottom, we could put in ## References. Okay. So we have references and then a list of our references. And that's pretty sweet, right? So we can do references, we can do tables, we can put figures into our R Markdown documents. So as I mentioned before, there's a lot of different output formats that we can get from R Markdown document, and this is all going to be set in that YAML header material. We've already seen how we can output HTML, as I mentioned we can output Markdown, we can output Word document files, PDF's, and many others. --- ## Output formats * [html](http://rmarkdown.rstudio.com/html_document_format.html) * [markdown](http://rmarkdown.rstudio.com/markdown_document_format.html) * [docx](http://rmarkdown.rstudio.com/word_document_format.html) * [pdf](http://rmarkdown.rstudio.com/pdf_document_format.html) * [Many others](http://rmarkdown.rstudio.com/lesson-9.html) <br> .center.alert[Each of these formats have many options that are constantly being added. Check the links to learn more] ??? Each of these formats have many, many, many options that are constantly being updated and added. So, I'd encourage you, if you're interested in that to click on those links and to go to the R Markdown site where they have really great documentation on how you can customize the output further. I'm going to give just a brief description of how we can do different things. --- ## html ``` --- title: Reproducing Kozich et al author: Joe Schmo date: "February 30, 2017" output: html_document bibliography: references.bib csl: mbio.csl #Get themes at https://github.com/citation-style-language/styles --- ``` <br> .center.alert[Possible to provide a CSS file, incorporate bootstrap, opengl, javascript, and other web-based tooling] ??? We've already seen with HTML using output: html_document. --- ## Markdown ``` --- title: Reproducing Kozich et al author: Joe Schmo date: "February 30, 2017" output: md_document bibliography: references.bib csl: mbio.csl #Get themes at https://github.com/citation-style-language/styles --- ``` <br> .center.alert[There are multiple flavors of markdown that you can select from within the different options] ??? With markdown, we can say output: md_document. --- ## docx / Word ``` --- title: Reproducing Kozich et al author: Joe Schmo date: "February 30, 2017" output: word_document bibliography: references.bib csl: mbio.csl #Get themes at https://github.com/citation-style-language/styles --- ``` <br> .center.alert[A useful-ish feature is the ability to apply a template to customize formatting. This can be rather limiting (e.g. no line numbers, problems with tables, bulleted lists, no forced line breaks). Alas, most people you work with will want a word file] ??? We can also do a word document, so word_document. It's a little bit beyond what I want to do right now, but we have the ability to generate a template to customize the formatting. It can be a little bit limiting. There might be ways to figure out line numbers, but it's not trivial, it problems with...some problems with table formatting, bulleted lists, and no forced line breaks. But at the same time, people you're working with are going to want a Word file, so what we generally do is we generate the Word file knowing that we're gonna send it to collaborators. We then manually format it to get the fonts and the spacing and everything the way we want it, and we send them the Word file. That maybe takes five minutes to kind of take what we get as a Word file out of Knitr to convert into what we want to send to our collaborators. --- ## pdf ``` --- title: Reproducing Kozich et al author: Joe Schmo date: "February 30, 2017" output: pdf_document bibliography: references.bib csl: mbio.csl #Get themes at https://github.com/citation-style-language/styles --- ``` <br> .center.alert[Must have a full installation of TeX on your system to render as PDF. This format offers the best looking output; however, there is a bit of a learning curve] ??? A PDF is also a powerful tool but you first have to have a full installation of TeX on your system to render it as a PDF. I think it offers the best possible output look but there's a bit of a learning curve and there's a variety of things that you're going to have to add using LaTeX. We have some help things built into the new project template that we're going to be using here in a minute. --- ## Combination ``` --- title: Reproducing Kozich et al author: Joe Schmo date: "February 30, 2017" output: pdf_document: default word_document: default md_document: default bibliography: references.bib csl: mbio.csl #Get themes at https://github.com/citation-style-language/styles --- ``` <br> ``` R -e "render('submission/practice.Rmd', output_format ='all')" ``` <br> .center.alert[I will often render a PDF and Word version for me and collaborators, respectively] ??? We can also generate output that's a combination of various types, PDF, Word, Markdown, whatever you want. Perhaps one of your collaborator wants a PDF, one wants a Word document, you know, go crazy. I'll often render a PDF and a Word version for me and my collaborators respectively because I like working with a PDF. I like the way it looks, it feels good to me. Of course, others want a Word document so they can do track changes. I still like to get a pen and kind of mark things up manually. --- ## `new_project` Rmd template * Open at the `manuscript.Rmd` file that is in `submission/` and render it as you did for `practice.Rmd` * Can you see how similar what we've been doing with `output: html_document` is to what is going on here? * Much of the special formatting (e.g. line numbers, line spacing, fonts, etc) are specified in header.tex, which is loaded in the YAML ??? So, we also have this new project Rmd template that was in there. You'll notice in the submission directory, there is a file called manuscript.Rmd. And we will render that as we did for practice.Rmd. --- ## Turning your document into a pdf * Copy your version of the "Scaling up" code and paragraph (without the YAML) into the "Results" section of `manuscript.Rmd` * Replace the figure caption in `manuscript.Rmd` with yours and your figure * Re-render ??? And so, hopefully, you'll see what's similar and what we're going to do is to copy what we had from that Scaling up paragraph into the results section of our manuscript, and we're going to go ahead and re-render what we had there. So again, if we do ls submission, we see down here that we have manuscript.Rmd and I should add that TeX comes preinstalled with this Amazon image. And like I said earlier, if you want to render it to PDF, you have to install TeX on your own and there's instructions on how to do that on the R Markdown website that's linked through those slide notes. So to open up manuscript.Rmd, and we see that there's some built-in YAML material here for you. There's also this set of Knitr settings for you that's already here. And we've got a running title, space, names for your authors. This is the cover page. You could customize this the way you want. There's an abstract, various sections of the paper. So, if you're writing a paper, you could take this template and you could populate all these sections. And then we've got figure legends, references, and so forth. So what we're going to do is I'm going to copy the material from practice into manuscript.Rmd. So do submission/practice, I'm going to use cat to output submission/practice.Rmd. And this will make it easier for me to copy and paste. So I'm going to grab this code chunk all the way down here, copy, nano submission/manuscript.Rmd, and I'm going to throw this in my results section. Okay. And then I'm going to take my figure 4 and my figure, copy that, and put this down at the bottom. And replace this here. Great. I'm going to save that. Back out. So again, what we've done is we've taken our results or Scaling up paragraph, put it into the results section of this manuscript.Rmd file. I'm going to save it, and now to render it, again, we can do R -e "render('submission/manuscript.Rmd')"We run that. It will generate manuscript.pdf for us. And we'll go to FileZilla. We see that we now have manuscript.pdf. We'll Download this. And we see we got out name of study. We can update this stuff if we wanted, of course. And so here, in our results and discussion Scaling up paragraph that has, again, the code we added in our inline code. And again, here's our figure. This was a PNG file. So its quality's pretty poor but, you know, we could update that and fix that as well. --- ## Closing note You can put code in your YAML and even [feed in parameters](http://rmarkdown.rstudio.com/developer_parameterized_reports.html) like a function ``` --- title: Reproducing Kozich et al author: Joe Schmo date: "`r Sys.time()`" output: html_document bibliography: references.bib csl: mbio.csl #Get themes at https://github.com/citation-style-language/styles --- ``` ??? All right? So again, we've seen how to output a HTML file as well as a PDF file, and looks pretty nice. We got line numbers, I've got that cover page that we could update and fix a bit to get it ready for submission. So as a closing note, we can also put code into our header material. So here's a way to put whatever today's date is in as the date, we're using `r Sys.time()` to get the time, the date that it will then go into the date for a report. We can also feed in parameters like a function treating the R Markdown document like a function where we might say, have grade reports or something like that we're doing for a class or some monthly report that we're doing for, kind of, updating a project. We can run that by feeding it specific data files to generate, kind of, reproducible reports where the underlying data is changing and it's then reflected in the output into the report. We've really done a lot today with R Markdown documents and thinking about how we can use R Markdown documents to make research reports as well as manuscripts that embed code into our science narrative. --- ## Exercises * .alert[Commit!] * Edit your "Scaling up" code and paragraph to automate the printing of another number or two * Generate a figure using the data in the shared file * If you have code in `code/utilities.R` or anoter similar code file, where would you run `source('code/utilities.R')`? ??? Of course, what we're going to want to do before we quit is to commit as well as log out of AWS. But before you do, I would really encourage you to edit your Scaling up code and paragraph further to automate the printing of another number or two from that paragraph. You might also see about generating a figure using the data in the shared file that we imported in that code chunk. Another question for you to think about, if you have code in a, say, a utility script called code utilities.R or something else like that, where would you run that source command to load the functions from that utilities.R file? I feel strongly that R Markdown and other literate programming tools are a huge step forward in improving the transparency and reproducibility of any analysis. Not only can someone re-run the code to generate the paper but they can see the code used to generate the numbers in the manuscript within the context of the actual text describing the relevance of the results. We saw R Markdown documents before when we looked at the meto paper published in the Journal of Microbiome. If you look back at that file, you'll notice they used R Markdown to generate what we might consider a research notebook type of document. That's a great way of walking people through an analysis in a less rigid manner than we typically see in a manuscript. My preference is to also make those type of documents but to use them more in a data exploration phase of a project and to use the tools we talked about today to prepare a polished manuscript that's ready for submission. Next time, we'll go all in and actually write our own write.paper function that allows us to start from an empty directory and end with a PDF version of our manuscript. We'll be using a tool called Make that will allow us to make our driver script more sophisticated and will allow us to restart the pipeline at any step in the workflow. Talk to you next time.