Automation with make
http://www.riffomonas.org/reproducible_research/make/ Press 'h' to open the help menu for interacting with the slides
Hi there, and welcome back. In the last tutorial, we discussed using literate programming tools like R Markdown to plunder narrative text for a manuscript or lab report with code to improve the transparency and reproducibility of our analyses.
The ongoing idea I've been developing throughout these tutorials is the idea of a write.paper command that will start with an empty directory and go all the way through our analyses to produce a final manuscript that is ready to submit. With material from the last tutorial, we are basically there, we could go ahead and add the necessary code to our driver script file and then we create our files.
One of the problems with that script though is that it's not very smart, consider for a moment that if instead of using the RDP taxonomy we instead use the SILVA or the Greengenes taxonomies. We'd have to either rerun the entire script or work out which files have the reference taxonomy as an upstream dependency. Today we'll learn to use a tool called Make, which keeps track of when a file was last updated as well as the other files that depend on it.
It's kind of a strange tool, but despite many efforts to recreate something like it, Make still persists as a dominant tool for building software packages. Inside of software packages we'll see how we can use it to keep track of a data analysis pipeline. Make will also allow us to deepen our thinking about the reproducibility of our analyses. We can write a function on Make to burn our project to the ground, and then we can use another function in Make to rebuild it.
We've seen this before with the analysis_driver script, and we'll reemphasize that principle today. Join me now in opening the slides for today's tutorial, which you can find within the reproducible research tutorial series at the riffomonas.org website.
Pop quiz
Noble's paper on organization of computational biology projects discusses far more than organization. How many of the principles that he mentioned can you remember?
Before we start talking about Make, I want to give you all a pop quiz that will jog your memory from a few sessions back, and help prime what we're going to be talking about today.
Noble's paper on organization of computational biology projects was something we've discussed a few times in previous sessions. I had this bulleted list of I think about six different things that Noble points out as being really important for organizing and going through a project. How many of those can you remember?
So maybe pause the video and scratch your head, maybe you look back through the previous notes if you have to, and then come back once you've got your list.
Pop quiz
Noble's paper on organization of computational biology projects discusses far more than organization. How many of the principles that he mentioned can you remember?
- "Record every operation that you perform"
- "Comment generously"
- "
Avoid[DO NOT] editing intermediate files by hand" - Use a driver script to centralize all processing and analysis
- "Use relative pathways [relative to the project root]"
- "Make the script restartable"
Before we start talking about Make, I want to give you all a pop quiz that will jog your memory from a few sessions back, and help prime what we're going to be talking about today.
Noble's paper on organization of computational biology projects was something we've discussed a few times in previous sessions. I had this bulleted list of I think about six different things that Noble points out as being really important for organizing and going through a project. How many of those can you remember?
So maybe pause the video and scratch your head, maybe you look back through the previous notes if you have to, and then come back once you've got your list.
This was the bulleted list that I've shown a few times to record every operation that you perform, and we talked about this in the driver script comment generously.
And again, this goes through all of our work, whether it's our driver script or our R files or our readme files, there's never enough documentation. We don't want to edit our intermediate files by hand. We want to be able to start the script, walk away and never have to touch anything.
Use a driver script to centralize all processing and analysis. Again, we want it to be automated, use relative pathways. And again, we've discussed we want to make this relative to the project root. Finally, the bullet point that we've kind of left to the side until now, until today's tutorial is to make the script restartable.
Learning goals
- Automate analyses with Makefiles
- Differentiate between rules, targets, and dependencies when building a Makefile
- Generate and use variables within a Makefile
- Take on a
make clean; makemindset to building a reproducible workflow
And so that's what we're going to talk about today in using Makefiles and how we can adapt our driver script file into a Makefile that will allow us to make that script restartable. We'll differentiate between different rules, targets, and dependencies when building a Makefile. We'll generate and use variables within a Makefile, and then we'll learn to take on make clean; make mindset to help us to build a reproducible workflow.
Driver script: Refresher from Noble
- "Record every operation that you perform"
- "Comment generously"
- "
Avoid[DO NOT] editing intermediate files by hand" - Use a driver script to centralize all processing and analysis
- "Use relative pathways [relative to the project root]"
- "Make the script restartable"
So again, really the focus of today's tutorial is taking this driver script where we centralized all our processing analysis, and making it a bit smarter, as I said earlier. We want to make the script restartable so that it can keep track of timestamps on when files were last modified or created, and what other files depend on those files.
Case study
You have finished analysis_driver.bash and are happy that you have automated your analysis. Consider the following scenarios...
- The mothur developers put out a new release of mothur
- There's a new version of the RDP classification database
- You decide to use the greengenes reference taxonomy
- You modify the color scheme for the ordination
- You've been persuaded to generate an animated version of the figure
So, a few case studies, imagine that we've finished making our analysis_driver.bash and we're happy that we've automated our analysis, but consider the following scenarios. So first, the Mothur developers have put out a new release of Mothur. It's always a possibility. We put one out last week actually. We're up to version 1.40 I think.
There might be a new version of the RDP classification database. Perhaps we decide to generate the Greengenes reference taxonomy. Or perhaps we have decided to change the coloring scheme for our ordination, instead of black and white, we want to make it, I don't know, orange and gray. Finally, perhaps we've been persuaded by RPI to generate an animated vision of our figure.
So for each of these, we might use our analysis_driver.bash file in different ways. We could start everything all over again from scratch, but that's going to take a while. But alternatively, there'd be a better way that we could restart up that script from the desired step.
The mothur developers put out a new release of mothur
So if we change the Mothur executable then perhaps we shouldn't have to change our downloads, and so forth. And so let's think about that initial example, that the Mothur developers put out a new release of Mothur. What all would we have to change?
The mothur developers put out a new release of mothur
- Regenerate
silva.seed.align(explicit dependency) - Rerun
get_good_seqs.batch(explicit dependency) - Rerun
get_error.batch(explicit dependency) - Rerun
get_shared_otus.batch(explicit dependency) - Rerun
get_nmds_data.batch(explicit dependency) - Rerun R code to generate figure (implicit dependency)
- Regenerate
manuscript.pdf(implicit dependency)
So if we change the Mothur executable then perhaps we shouldn't have to change our downloads, and so forth. And so let's think about that initial example, that the Mothur developers put out a new release of Mothur. What all would we have to change?
And so if we think about the various steps in the pipeline, well, we'd have to regenerate silva.seed.align, this explicitly depends on Mothur. We'd have to rerun get_good_seqs.batch, get_error.batch, get_shared_otus batch, get_nmds batch, and we have to rerun our R code and generate the figure. This is what we might consider an implicit dependency, so to run the R code we don't run Mothur, we don't use Mothur, but we use files generated by Mothur.
So the output of get_shared_otus.batch goes into get_nmds batch. And then that data that was generated by Mothur is then run in R. And then finally, we'd have to regenerate manuscript.pdf. Again, Mothur isn't used to generate manuscript.pdf, but information that goes into manuscript.pdf was at one point generated by Mothur.
There's a new version of the RDP classification database
Let's think about if there's a new version of the RDP classifier.
There's a new version of the RDP classification database
- Download new version of database (explicit dependency)
- Rerun
get_good_seqs.batch(explicit dependency) - Rerun
get_error.batch(implicit dependency) - Rerun
get_shared_otus.batch(implicit dependency) - Rerun
get_nmds_data.batch(implicit dependency) - Rerun R code to generate figure (implicit dependency)
- Regenerate
manuscript.pdf(implicit dependency)
Let's think about if there's a new version of the RDP classifier.
Well, we'd have to download the new version of the database, so that's an explicit dependency. We then have to rerun get_good_seqs.batch, because there's a step in there were remove sequences that are, say, chloroplasts or mitochondria, we also might update our taxonomy assignments for each OTU, we'd have to then rerun get_error.batch, because again in there is how we…it's affected by how we've perhaps removed sequences based on their taxonomy. We'd have to rerun then get_shared_otus.batch, and get_nmds_data.batch. So each of these last three commands are again implicit dependencies, because data that's generated upstream of that is being affected by having a new reference taxonomy, and again, we'd have to regenerate our R code and our figure and rerun manuscript.pdf.
You decide to use the greengenes reference taxonomy
So say we wanted to update to use the Greengenes reference taxonomy.
You decide to use the greengenes reference taxonomy
- Download new version of database (explicit dependency)
- Rerun
get_good_seqs.batch(explicit dependency) - Rerun
get_error.batch(implicit dependency) - Rerun
get_shared_otus.batch(implicit dependency) - Rerun
get_nmds_data.batch(implicit dependency) - Rerun R code to generate figure (implicit dependency)
- Regenerate
manuscript.pdf(implicit dependency)
So say we wanted to update to use the Greengenes reference taxonomy.
Again, it's all the same things as changing the version of the RDP reference, but instead we're going to be using the Greengenes reference.
You modify the color scheme for the ordination
Say we want to modify the coloring scheme of our ordination.
You modify the color scheme for the ordination
- Rerun R code to generate figure (explicit dependency)
- Regenerate
manuscript.pdf(implicit dependency)
Say we want to modify the coloring scheme of our ordination.
Well, that was a kind of a later step in our R script, we wouldn't have to rerun all those previous Mothur commands because the Mothur commands don't depend on what we use for a color scheme, so it would be considerably simpler to generate a new version of the figure with a different color scheme than to say change the reference taxonomy that we're using.
You've been persuaded to generate an animated version of the figure
Say we've been persuaded to generate an animated version of the figure.
You've been persuaded to generate an animated version of the figure
- Write and run new R script (explicit dependency)
- Regenerate
manuscript.pdf(maybe)
Say we've been persuaded to generate an animated version of the figure.
Well. to do that we'd have to write and run a new R script, and then we might have to regenerate manuscript.pdf. So we might not have to do that because maybe that figure wouldn't go into the manuscript, because there aren't too many animated figures in a manuscript.
Dependency hell
- All of these dependencies are difficult to keep track of
- The situation only worsens as the analysis becomes more complicated
- You could use
ifstatements that check time stamps usingbash
So this is what we call dependency hell, all of these dependencies are difficult to keep track of, and yes, you could do it by hand or by tracing your finger back through the files, but it gets really complicated and this is just hellish. The situation only gets worse as the analysis becomes more complicated. You can imagine these dependency as being kind of this like tangled web.
We could also, we could take our analysis_driver script, we could make it smarter using if statements and checking timestamps using various bash commands. But that's all kind of a pain in the neck. So what are we going to use instead?
Dependency hell
- All of these dependencies are difficult to keep track of
- The situation only worsens as the analysis becomes more complicated
- You could use
ifstatements that check time stamps usingbash
Make to the rescue!
So this is what we call dependency hell, all of these dependencies are difficult to keep track of, and yes, you could do it by hand or by tracing your finger back through the files, but it gets really complicated and this is just hellish. The situation only gets worse as the analysis becomes more complicated. You can imagine these dependency as being kind of this like tangled web.
We could also, we could take our analysis_driver script, we could make it smarter using if statements and checking timestamps using various bash commands. But that's all kind of a pain in the neck. So what are we going to use instead?
We can use Make.
Make
- Developed by Stuart Feldman in 1977 as a Bell Labs summer intern
- The goal was to have a tool for compiling software
- People generally hate make – hence the knock offs
- Recognize that using it for data analysis wasn't the original intent
- Hard to do much better!
Make is a tool that was developed in the 1970s by Stuart Feldman when he was a summer intern at Bell Labs. I think Bell Labs is pretty remarkable, and I suspect Stuart Feldman was a pretty remarkable individual to have a tool persist for the last 40 years from a summer internship. I don't know what you were doing for your summer internships, but I certainly wasn't making a tool that would last that long. So the goal was to have a tool for compiling software. People generally hate Make. It's really, it can be frustrating to use, I'll try to make it as simple as possible. So there's a variety of knockoffs that are trying to do what Make does but slightly easier to use perhaps. But again, it's been going for 40 years and none of the knockoffs have taken over, so it's clear that it's hard to do much better than the original. So then the primary use case is from software engineering, where we want to recompile software but we don't want to have to do the entire thing.
Primary use case
Don’t want to recompile an entire program with a complex codebase for every change, so only compile what changed and what depends on that change
To make Mothur, we might have 200 files that get compiled to make that one Mothur file. Well, I don't want to have to recompile everything that's in there if I'm only changing one file, I only want to change the things that depend on that file.
Primary use case
Don’t want to recompile an entire program with a complex codebase for every change, so only compile what changed and what depends on that change
Our use case
Don't want to re-run or re-download files that require a lot of time to generate, so only rebuild files that depend on that change
To make Mothur, we might have 200 files that get compiled to make that one Mothur file. Well, I don't want to have to recompile everything that's in there if I'm only changing one file, I only want to change the things that depend on that file.
Our use case with a data analysis is that I perhaps don't want to rerun or redownload files that require a lot of time to generate, so I only want to rebuild the files that depend on that change.
Motivation
- I will repeat various steps in an analysis multiple times
- The end product is a data-heavy paper written in Rmarkdown
- Each step of the analysis may be slow
- There are a lot of steps
- Hard to keep track of all of the dependencies
- Analysis run on an HPC, so it needs to be scripted
- Want to make it possible for others (including me!) to replicate what I’ve done
So my motivation is that I'm going to repeat various steps and analysis many times. I might be tweaking different parameters, I might do the thing where I change my reference taxonomy. The end product is going to be a data-heavy paper written in R Markdown as we discussed in the last tutorial.
There's going to be steps in the analysis that are slow, I don't want to have to do a lot of hand-holding, and there's going to be a lot of these steps. And so I want it to be as automated as possible. And because the projects get complicated, it's frequently hard to keep track of all the dependencies. Also, when I do this, I'm going to run it on a high performance computer like Flux here at University of Michigan, or as we've been doing on Amazon's Cloud.
So this needs to be scripted as much as possible so I can use something like tmux or a scheduler to fire off the job and walk away. Also I want to make it possible for others, including me, to replicate what I've done.
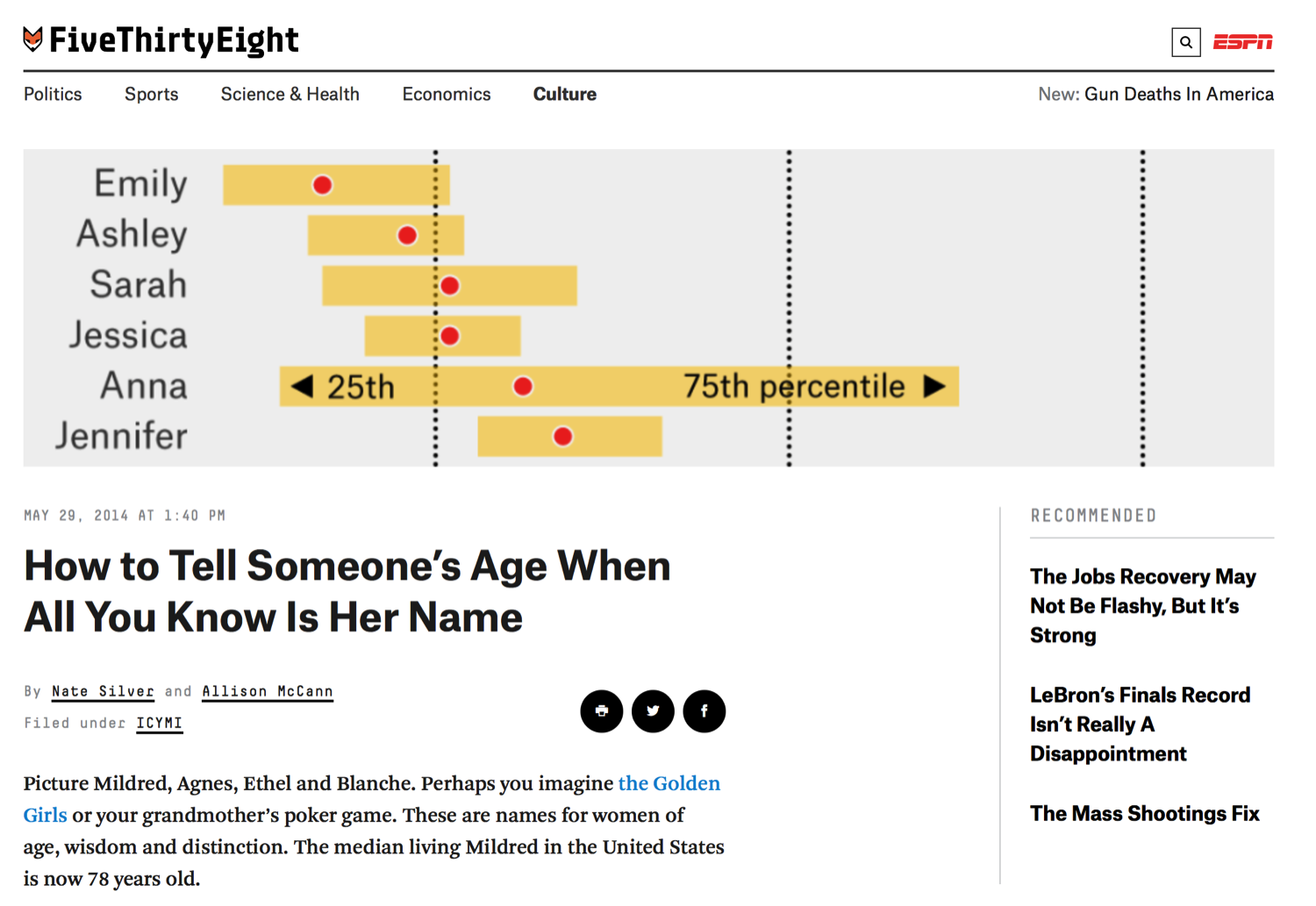
So to get us introduced to Make, we're going to take on a fun example stolen from fivethirtyeight.com, where they have a story about predicting the age of individuals based on their name.
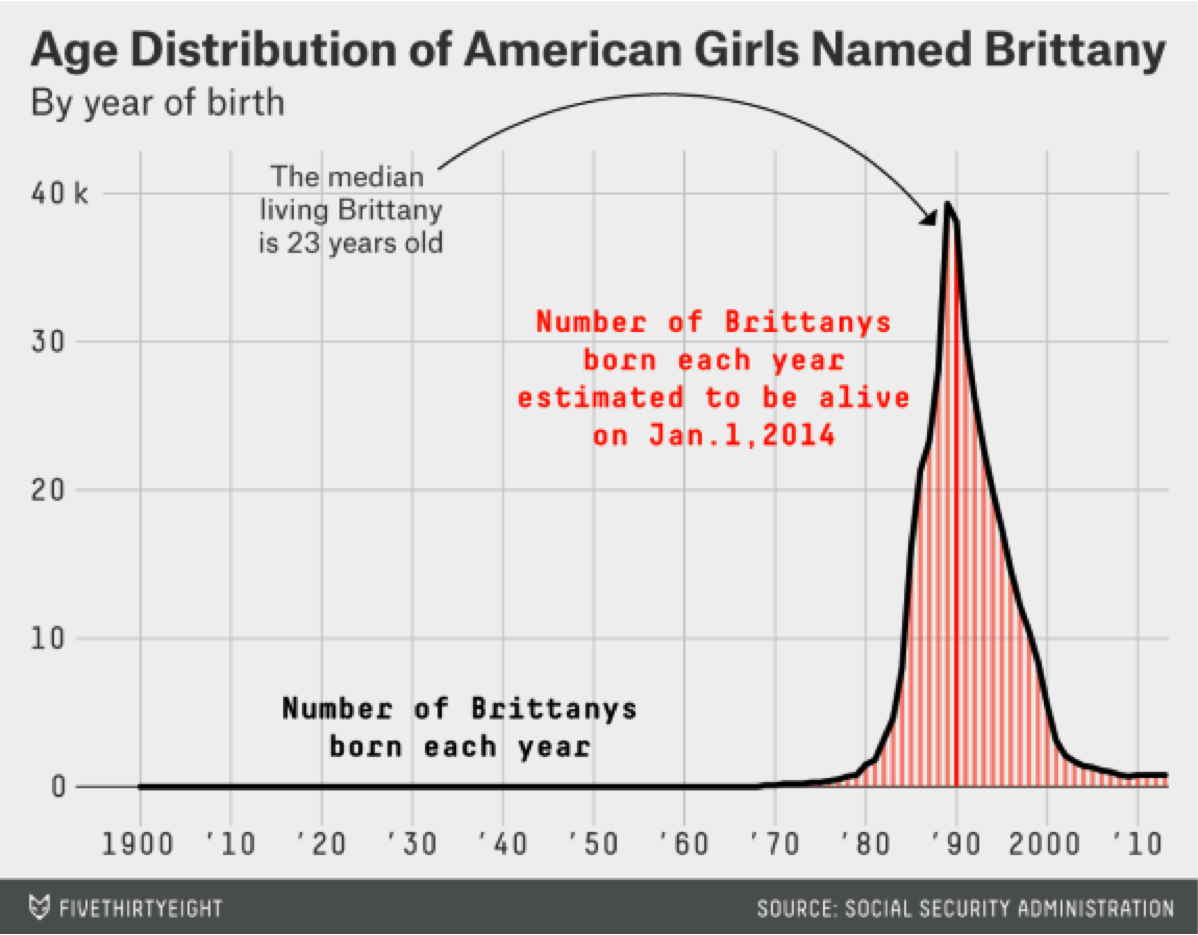
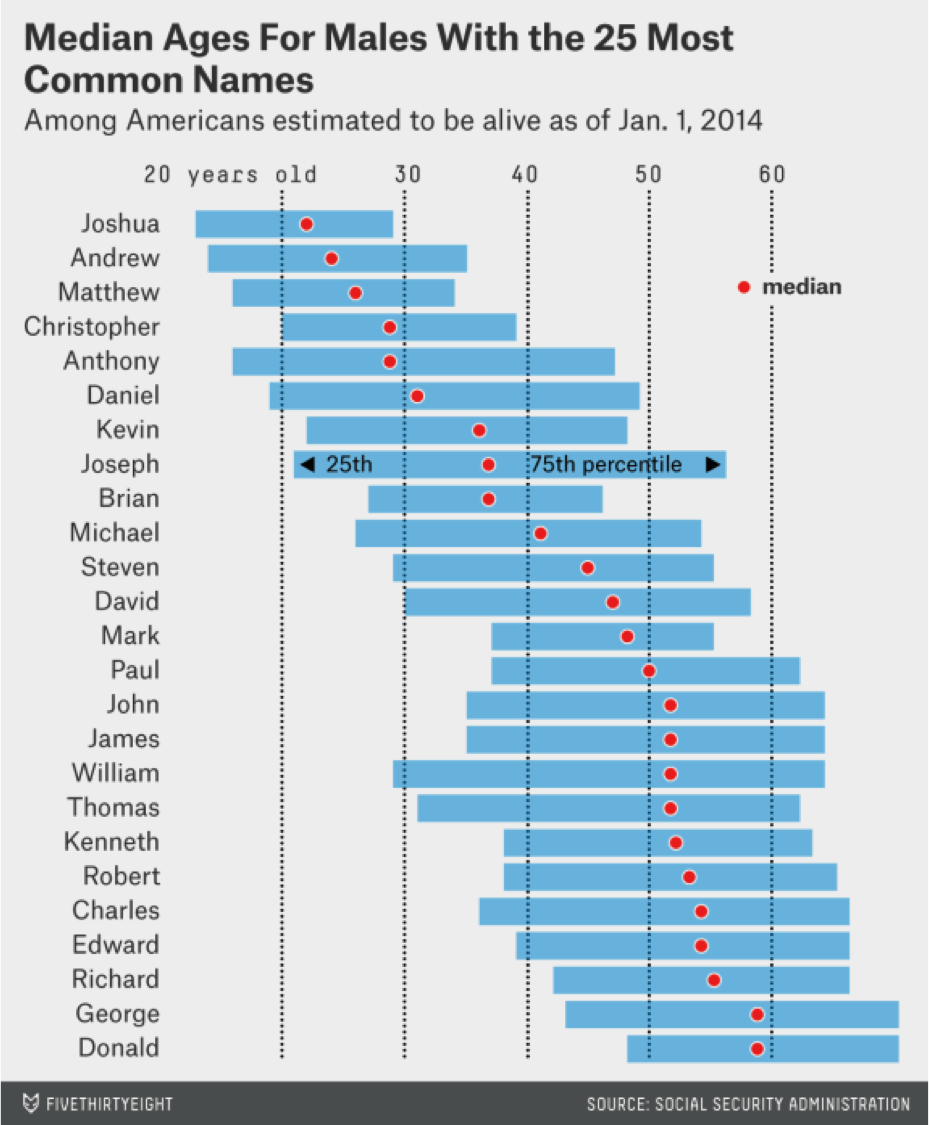
So if I told you my name was Patrick, can you predict how old I am just based on my name? And so here's an example of them doing that with American girls named Brittany by year of birth, and so if somebody's name was Brittany you might expect that they were born around 1990. Here's the distribution from their paper, among the 25 most common boys names, Patrick doesn't show up here. But if you had a boy, let's say, named Anthony, you would expect that the median is about 28 years old.
Reactions
- What about my kids’ names?
- This would be cool as a Shiny app
- This was done in 2013, what about 2017?
- What if we had similar data from Canada?
- Can I replicate their analysis?
So my reaction to reading this, I love baby names, I have a bunch of kids, and so I'm always wondering how my kids fit into this. And so I wondered, "What about my kids' names? Would I predict their ages based on their names? And what about my own name? What about my wife's name?"
And so if you know about R, there is this package called Shiny which I could imagine this analysis being really cool as a Shiny app where you can plug in any name and get something out. This analysis was done in 2013. Well, what about if we did it today, 2017, 2018? It was also for kids in the United States, but what about kids from Canada?
And then ultimately, can I replicate their analysis? I want to understand how they did their analysis, and can I get that get their work to be replicated?
Reactions
- What about my kids’ names?
- This would be cool as a Shiny app
- This was done in 2013, what about 2017?
- What if we had similar data from Canada?
- Can I replicate their analysis?
This is a great opportunity for thinking about reproducible research
So my reaction to reading this, I love baby names, I have a bunch of kids, and so I'm always wondering how my kids fit into this. And so I wondered, "What about my kids' names? Would I predict their ages based on their names? And what about my own name? What about my wife's name?"
And so if you know about R, there is this package called Shiny which I could imagine this analysis being really cool as a Shiny app where you can plug in any name and get something out. This analysis was done in 2013. Well, what about if we did it today, 2017, 2018? It was also for kids in the United States, but what about kids from Canada?
And then ultimately, can I replicate their analysis? I want to understand how they did their analysis, and can I get that get their work to be replicated?
So this is a great opportunity for thinking about reproducible research.
I really admire FiveThirtyEight because they've done a great job of making a lot of their data and code available on GitHub. So it's great that they're doing open science, as you can tell if you look at their data repository, it's kind of a garbage can, it's all there, you just have to dig a while, it's not really well organized. And the script files in here, this most-common-name.R file doesn't actually generate the figures from the paper, so that's a bit frustrating.
So I did it for them...
$ cd ~$ git clone https://github.com/riffomonas/make_tutorial.git$ cd make_tutorial$ bash baby_name_analysis.bash- My GitHub repository
- Note that this is being driven by a single bash file
- It is a mixture of bash commands (e.g. curl, unzip) and R calls
- Note: you will need to install the
zooR package
So I did it for them, and this is what we're going to work on. And so the idea is that we're going to be able to go into our home directory, run git clone to download my version of their analysis as a analysis driver script. We're going to move into that directory and then we're going to run that and generate the files.
So this is the idea, that we have this driver script, and it does the analysis. And so what we're going to do is we're going to convert this baby_name_analysis.bash file into a Makefile as an illustration of then what we're going to do with our Kozich data. So we're going to go ahead and log into AWS to do this analysis.
So we're logged in. I'm going to go ahead and do git clone https. Great.
I cloned down cd into my make_tutorial, ls. And again, if we wanted to, we could look through here.
Let's look at our nano baby_name_analysis.bash. And you can see I've hopefully commented generously through here. I've listed my dependency, what things are being produced, my R code, the various commands I'm running to do all this.
I can log out of nano. If I look in data, I've got my processed and raw. There's nothing in processed. There's one CSV in my raw directory, and this is a file that indicates the number of people that were alive in 2016 I guess per 100,000.
So, the other thing that I have in my notes is that my code requires the zoo package. So if I go into R and I do library(zoo), there's no package called zoo, so I need to install the zoo package. So I'll do install.packages("zoo").
So that's installed now, so I can quit. And clear my screen, and I should be able to run this by running the bash command. So I'm going to go ahead and do tmux, because I forget how long exactly this takes.
And we can then do bash baby_name_analysis.bash, hit Enter, and sit back and let it roll. So, that was pretty quick. Let's go ahead and open up FileZilla. And I'm going to go into make_tutorial.
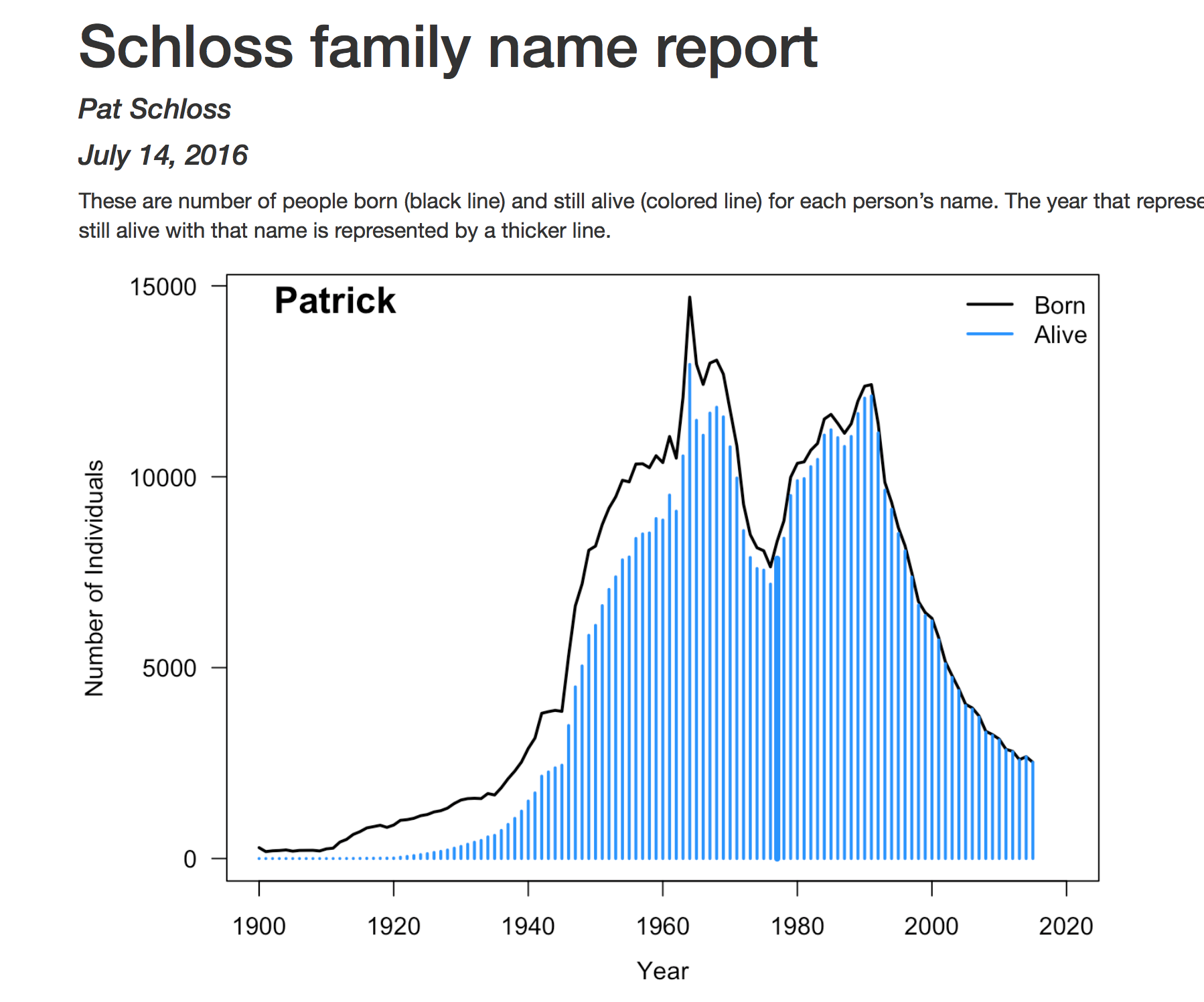
And I've got family_report.html. And voilà.
So, I did this for my kids' names, Patrick, Joe, John, Jacob, Peter, Ruth, Mary, Martha. I have another kid that I forgot to add, Simon, maybe we can do that as part of this. And so you can see the number of people that were born in each year named Patrick, as well as the number of people today or 2016 that were alive.
So I have a son named Patrick, my name is Patrick. This line here, 76, right about is when I was born, and so we can see that Jacob was… This solid, blue line is the median, Jacob was born in the 2010s, so that's kind of close.
And, oh I love this data, I think it's kind of cool. Martha was born four years ago, so this gets her quite a bit wrong. And so you can see my version of their plot here, where you have the ages of the different children along with the inner quartile range for each child and the median year or age I guess that we would have expected them to be born.
So that's pretty cool. Let's think about how we would add Simon to this analysis, and what that would do to the overall flow of our project.
#!/usr/bin/env bash# Prepare annual baby name data report.# Assumes:# * the user is connected to the internet# * has R installed in their PATH# * has rmarkdown package installed# Depends on: website data at https://www.ssa.gov/oact/babynames/names.zip# Produces: slew of files named yob????.txt and a pdf filewget --no-check-certificate -P data/raw/ https://www.ssa.gov/oact/babynames/names.zipunzip -d data/raw/ data/raw/names.zip# Concatenate the annual baby name data# Depends on: data/raw/yob????.txt files# code/concatenate_files.R# Producees: data/processed/all_names.csvR -e "source('code/concatenate_files.R')"# Fills in missing data from annual survivorship data# Depends on: data/raw/alive_2016_per_100k.csv# code/interpolate_mortality.R# Produces: data/processed/alive_2016_annual.csvR -e "source('code/interpolate_mortality.R')"# Generate counts of total and living people with each name# Depends on: data/processed/alive_2016_annual.csv# data/processed/all_names.csv# code/get_total_and_living_name_counts.R# Produces: data/processed/total_and_living_name_counts.csvR -e "source('code/get_total_and_living_name_counts.R')"# Renders an Rmarkdown file that creates various plots and# provides an entertaining color commentary# Depends on: data/processed/total_and_living_name_counts.csv# code/plot_functions.R# Produces: family_report.htmlR -e "library(rmarkdown); render('family_report.Rmd')"So again, this is the driver script file, where again we have good documentation and comments, and then we have a wget which pulls the data down from the Social Security office, we unzip that and we move the data to the right place.
This is similar to what we've already done with our Kozich data. We then run an R script that concatenates the annual baby name data. We then fill in the missing data from the annual survivorship data to interpolate the mortality, because I think it came out every five years or every couple years, and so we need to interpolate the intervening years. We then generate counts of total and living people with each name using an R script, and then we run our R Markdown to render the family_report.Rmd to generate that final HTML file.
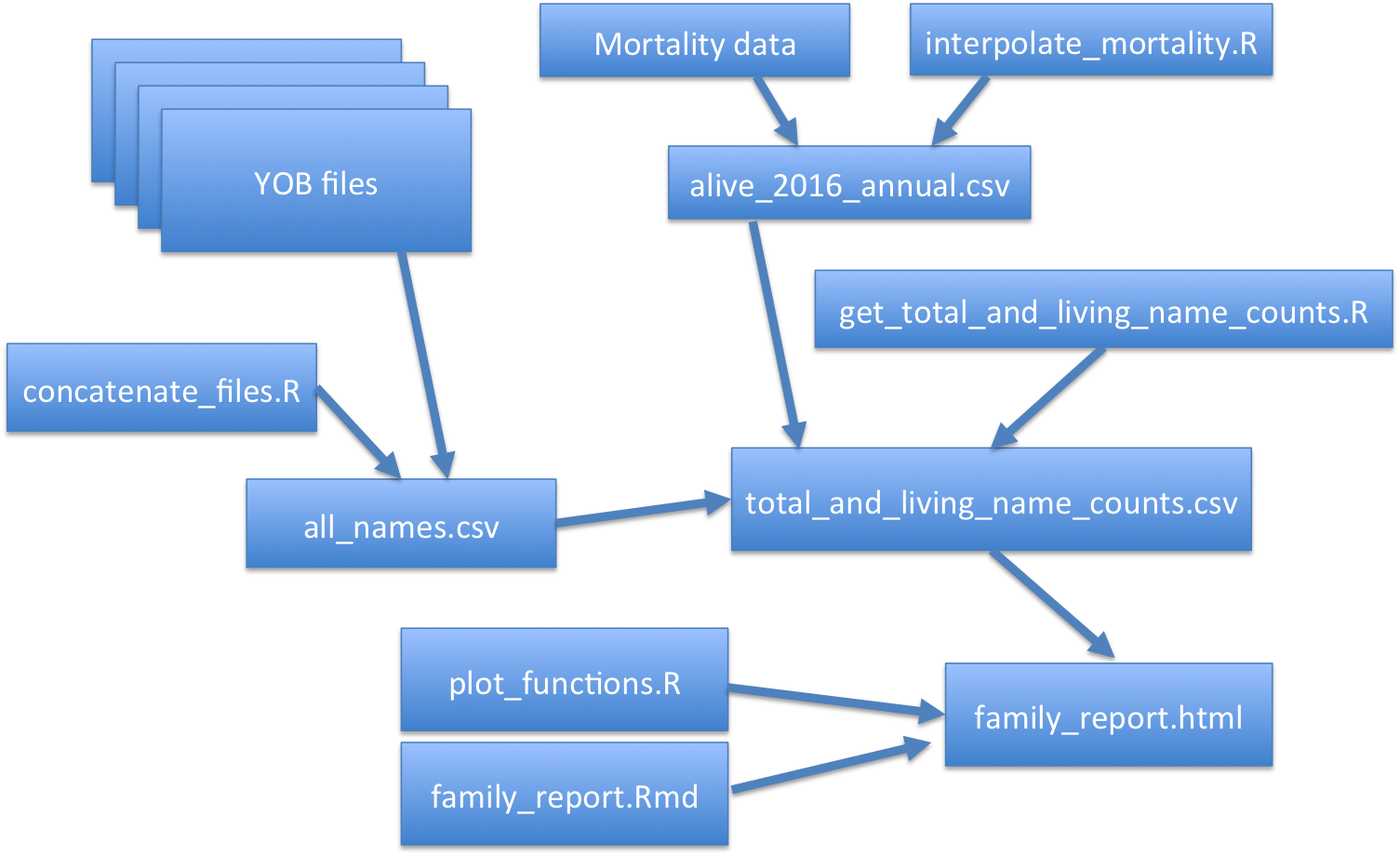
So if we look at this as a diagram, we can see that the goal is a family_report.html file, but to generate this depends on family_report.Rmd and plot_functions.R. It also, so this is code, it also depends on data, total_and_living_name_counts.csv.
And so this then is dependent on an R script and other data file which is also dependent on that mortality data that we already saw in the data/raw directory, as well as another R script. And it's also dependent on other data. And so we can see that if we change one of these, if I say change or delete all_name.csv, I'm going to have to regenerate this because that is going to be needed here and it's going to be needed here.
Whereas if I just generate plot_functions.R, I don't have to worry about all this other stuff, I only need to worry about regenerating family_report.html. If I say change the code in interpolate_mortality.R, then everything on this side of the network is going to need to be updated.
What I want...
$ git clone https://github.com/pschloss/make_tutorial.git$ cd make_tutorial$ make family_report.htmlAnd it just works!
So, what we'd like to do is similar to what we had done previously. I want to be able to clone the repository, do cd make_tutorial, and then write make family_report.html, and it just works.
Makefile structure
A makefile is a series of rules that contain commands for processing the dependencies to create the targets
data/processed/all_names.csv : code/concatenate_files.R $(YOBTXT) R -e "source('code/concatenate_files.R')"And so as we've discussed, a Makefile is a series of rules as they're called, and these rules contain commands for processing the dependency to create what are called the targets.
Makefile structure
A makefile is a series of rules that contain commands for processing the dependencies to create the targets
data/processed/all_names.csv : code/concatenate_files.R $(YOBTXT) R -e "source('code/concatenate_files.R')"- Target: stuff to the left of the
:(data/processed/all_names.csv) - Dependencies: stuff to the right of the
:(code/concatenate_files.R) - Commands:
R -e "source('code/concatenate_files.R')"
And so as we've discussed, a Makefile is a series of rules as they're called, and these rules contain commands for processing the dependency to create what are called the targets.
So we have commands, we have dependencies, and we have targets. So the target is the stuff on the left of the colon, so the target here is data/processed/all_names. The dependencies is all the stuff to the right, so code/concatenate_files.R, and then there's $(YOBTXT).
Important point!
The second line is inset by a TAB, not spaces
data/processed/all_names.csv : code/concatenate_files.R $(YOBTXT) R -e "source('code/concatenate_files.R')"These are the dependencies. The commands then are what follows on the following line, set off by a tab. So this R –e "source…" blah-blah-blah, that is the command. And so it's important to note, cannot be emphasized enough, I make this mistake frequently.
The second line is set off by a tab, not spaces. So this is a tab character, not spaces. And you will get an error if you put spaces in there, it will complain.
What does this rule allow us to do?
I can generate data/processed/all_names.csv with...
make data/processed/all_names.csvSo what does this rule allow us to do? Well, I can generate data/processed/all_names.csv with this Make command. So what we're going to do is we're going to convert our analysis_driver.batch file into a Makefile.
Let's convert to a Makefile
# Renders an Rmarkdown file that creates various plots and# provides an entertaining color commentary# Depends on: data/processed/total_and_living_name_counts.csv# code/plot_functions.R# Produces: family_report.htmlR -e "library(rmarkdown); render('family_report.Rmd')"
How would you write this as a rule?
Note that you can extend lines with a \
So looking at this, how would we write this as a rule? And note that if you have really long lines, you can extend the lines with a backslash. Well, we could take this and say that the target is family_report.html. The dependencies are family_report.Rmd, code/plot_functions.R, and data/processed/total_and_living_names.csv.
And then the command is down here. So I'm going to do some copying and pasting. And so I'm going to copy this, I'm going to create or open my Makefile, so I do nano Makefile. It's an empty file, I can paste it into here.
This now is my Makefile. So if I save that, I can do make family_report.html, runs. Aah, missing separator. That is the error when there are tabs. So if I go back in here, ah, yes, so you can see I'm inching over. So I need to put a tab in there, I exit, make family_report, it says it's up to date.
Let's convert to a Makefile
# Renders an Rmarkdown file that creates various plots and# provides an entertaining color commentary# Depends on: data/processed/total_and_living_name_counts.csv# code/plot_functions.R# Produces: family_report.htmlR -e "library(rmarkdown); render('family_report.Rmd')"
How would you write this as a rule?
Note that you can extend lines with a \
# Renders an Rmarkdown file that creates various plots and# provides an entertaining color commentary# Depends on: data/processed/total_and_living_name_counts.csv# code/plot_functions.R# Produces: family_report.htmlfamily_report.html : family_report.Rmd\ code/plot_functions.R\ data/processed/total_and_living_name_counts.csv R -e "library(rmarkdown); render('family_report.Rmd')"Copy and paste this into Makefile
So looking at this, how would we write this as a rule? And note that if you have really long lines, you can extend the lines with a backslash. Well, we could take this and say that the target is family_report.html. The dependencies are family_report.Rmd, code/plot_functions.R, and data/processed/total_and_living_names.csv.
And then the command is down here. So I'm going to do some copying and pasting. And so I'm going to copy this, I'm going to create or open my Makefile, so I do nano Makefile. It's an empty file, I can paste it into here.
This now is my Makefile. So if I save that, I can do make family_report.html, runs. Aah, missing separator. That is the error when there are tabs. So if I go back in here, ah, yes, so you can see I'm inching over. So I need to put a tab in there, I exit, make family_report, it says it's up to date.
So if I were to go in and do nano family_report.Rmd. And let's say I say, "Schloss family name report (without poor Simon!)". So, I have now updated one of the dependencies for family_report.html.
Try it out
$ make family_report.htmlmake: `family_report.html' is up to date.I can again do make family_report.html, and it renders that step. It will run that step, and it now makes family_report.html.
Next...
# Generate counts of total and living people with each name# Depends on: data/processed/alive_2016_annual.csv# data/processed/all_names.csv# code/get_total_and_living_name_counts.R# Produces: data/processed/total_and_living_name_counts.csvR -e "source('code/get_total_and_living_name_counts.R')"
How would you write this as a rule?
So the next rule in our analysis_driver.batch file, generates a file that contains the counts of the total and living people with each name, and it depends on these three files, two sets of data, and an R script.
And it then produces data/processed/total_and_living_name_counts. Go ahead and pause the video, and see if you can convert these dependencies, target, and code as a rule for Make.
Next...
# Generate counts of total and living people with each name# Depends on: data/processed/alive_2016_annual.csv# data/processed/all_names.csv# code/get_total_and_living_name_counts.R# Produces: data/processed/total_and_living_name_counts.csvR -e "source('code/get_total_and_living_name_counts.R')"
How would you write this as a rule?
# Generate counts of total and living people with each name# Depends on: data/processed/alive_2016_annual.csv# data/processed/all_names.csv# code/get_total_and_living_name_counts.R# Produces: data/processed/total_and_living_name_counts.csvdata/processed/total_and_living_name_counts.csv : code/get_total_and_living_name_counts.R\ data/processed/alive_2016_annual.csv\ data/processed/all_names.csv R -e "source('code/get_total_and_living_name_counts.R')"So the next rule in our analysis_driver.batch file, generates a file that contains the counts of the total and living people with each name, and it depends on these three files, two sets of data, and an R script.
And it then produces data/processed/total_and_living_name_counts. Go ahead and pause the video, and see if you can convert these dependencies, target, and code as a rule for Make.
So again, what we might expect is that on the left side of the colon, we're going to have data/processed/ total_and_living_name_counts.csv.
We're going to have a colon, and then we're going to list these three files. And then on the next line our command is going to be this R call. And sure enough, that's what we see here.
Next...
# Generate counts of total and living people with each name# Depends on: data/processed/alive_2016_annual.csv# data/processed/all_names.csv# code/get_total_and_living_name_counts.R# Produces: data/processed/total_and_living_name_counts.csvR -e "source('code/get_total_and_living_name_counts.R')"
How would you write this as a rule?
# Generate counts of total and living people with each name# Depends on: data/processed/alive_2016_annual.csv# data/processed/all_names.csv# code/get_total_and_living_name_counts.R# Produces: data/processed/total_and_living_name_counts.csvdata/processed/total_and_living_name_counts.csv : code/get_total_and_living_name_counts.R\ data/processed/alive_2016_annual.csv\ data/processed/all_names.csv R -e "source('code/get_total_and_living_name_counts.R')"
Paste it into the head of Makefile and build the target
So the next rule in our analysis_driver.batch file, generates a file that contains the counts of the total and living people with each name, and it depends on these three files, two sets of data, and an R script.
And it then produces data/processed/total_and_living_name_counts. Go ahead and pause the video, and see if you can convert these dependencies, target, and code as a rule for Make.
So again, what we might expect is that on the left side of the colon, we're going to have data/processed/ total_and_living_name_counts.csv.
We're going to have a colon, and then we're going to list these three files. And then on the next line our command is going to be this R call. And sure enough, that's what we see here.
I'm going to copy this, and paste it into my Makefile. And this is upstream of family_report.html, so I'm going to put it above that. I don't think the order necessarily matters. And I'm going to put it, replace those spaces on the command line with a tab. I don't think the order necessarily matters except for my own readability. And speaking of readability, I'm going to make this a little bit wider so that the lines don't have funky breaks, that's one of the problems with these really long filenames.
So we can save this, quit out, and I can then do make data/processed/ total_and_living_name_counts.csv. And it says it's up to date, we're good to go. So the dependencies that we have in here are these three files, so if we were to change get_total_and_living_name_counts.R, it would re-run this, and it would also rerun our render for the R Markdown file.
So let's give that a shot. So if we do nano code/get_total_and _living_name_counts.R, I'm going to remove this note to myself just to have something to edit. Save that, quit out. So if I now do make family_report.html, clear the screen so it's easier to see, we'll see that that first step, it went by pretty quick, the first step was to regenerate that file and then to regenerate family_report.html.
Getting the hang of this?
# Fills in missing data from annual survivorship data# Depends on: data/raw/alive_2016_per_100k.csv# code/interpolate_mortality.R# Produces: data/processed/alive_2016_annual.csvR -e "source('code/interpolate_mortality.R')"
How would you write this as a rule?
So again, it's keeping track of these dependencies for us which is really powerful and really nice. So we're getting the hang of this. Here's a third one, and what I'd like you to do is go ahead and pause the video, make the rule to generate the data/processed/alive_2016_annual.csv file. Go ahead and paste this into your Makefile and see if you can get it to work. So again, convert what we had in analysis_driver.batch, put it into your makefile, make sure it's properly formatted, and then go ahead and run the make command, maybe change interpolate_mortality.R, and make sure that everything works the way you would expect it to.
Getting the hang of this?
# Fills in missing data from annual survivorship data# Depends on: data/raw/alive_2016_per_100k.csv# code/interpolate_mortality.R# Produces: data/processed/alive_2016_annual.csvR -e "source('code/interpolate_mortality.R')"
How would you write this as a rule?
# Fills in missing data from annual survivorship data# Depends on: data/raw/alive_2016_per_100k.csv# code/interpolate_mortality.R# Produces: data/processed/alive_2016_annual.csvdata/processed/alive_2016_annual.csv : data/raw/alive_2016_per_100k.csv\ code/interpolate_mortality.R R -e "source('code/interpolate_mortality.R')"
Paste it into the head of Makefile and build the target
So again, it's keeping track of these dependencies for us which is really powerful and really nice. So we're getting the hang of this. Here's a third one, and what I'd like you to do is go ahead and pause the video, make the rule to generate the data/processed/alive_2016_annual.csv file. Go ahead and paste this into your Makefile and see if you can get it to work. So again, convert what we had in analysis_driver.batch, put it into your makefile, make sure it's properly formatted, and then go ahead and run the make command, maybe change interpolate_mortality.R, and make sure that everything works the way you would expect it to.
So hopefully you were able to add this rule in to generate the alive_annual_2016.csv file, if we save it, quit out, we do make family_report.html, everything is up to date. You can get a long ways with using make by making these relatively simple rules, where you, again, define your target, your dependencies, and the command you want to run.
The next step is a little harder
- We need to generate a large number of dependencies
The next step gets a little bit harder because we need to generate a list of a large number of dependencies, because we're going to get data from all sorts of years, from 1900 to the year 2015. And we're going to require some bash to do that, and some of the built-in functions for Make that will allow us to generate filenames.
The next step is a little harder
We need to generate a large number of dependencies
makehas built in functions that allow you to construct dependency names
YEARS=$(shell seq 1900 2015)YOB=$(addprefix data/raw/yob,$(YEARS))YOBTXT=$(addsuffix .txt,$(YOB))The next step gets a little bit harder because we need to generate a list of a large number of dependencies, because we're going to get data from all sorts of years, from 1900 to the year 2015. And we're going to require some bash to do that, and some of the built-in functions for Make that will allow us to generate filenames.
To break down these three lines for us, shall seek which we see here, generates numbers between 1900 and 2015. So, it generates a vector of numbers between 1900 and 2015. So if we're to look at the value of years it'd be 1901, 1902, 1903, all the way up to 2015.
The next step is a little harder
We need to generate a large number of dependencies
makehas built in functions that allow you to construct dependency names
YEARS=$(shell seq 1900 2015)YOB=$(addprefix data/raw/yob,$(YEARS))YOBTXT=$(addsuffix .txt,$(YOB))shell seqgenerates a numbers between 1900 & 2015addprefixadds prefix (data/raw/yob) to each value ofYEARS$(YEARS)is how we get the values of theYEARSarrayaddsuffixadds suffix (.txt) to each value ofYOBTXT
The next step gets a little bit harder because we need to generate a list of a large number of dependencies, because we're going to get data from all sorts of years, from 1900 to the year 2015. And we're going to require some bash to do that, and some of the built-in functions for Make that will allow us to generate filenames.
To break down these three lines for us, shall seek which we see here, generates numbers between 1900 and 2015. So, it generates a vector of numbers between 1900 and 2015. So if we're to look at the value of years it'd be 1901, 1902, 1903, all the way up to 2015.
We could then have year of birth, YOB as file, where we will add prefix data for where we will add raw data, raw YOB to each of the years. If we look at the data/raw, we see that we've downloaded from names.zip all of these files of yob1880 and so forth.
We're starting at 1900 because I think the data before 1900 are kind of spotty. You also see that we have 2016.txt, I think the data have been released since we initially, I developed this tutorial. So what we're trying to do is generate a path that's data/raw/yob1902.txt.
So you can see what we're trying to do is build that path that goes data/raw/year-of-birth, yob, and then the number. And then we can use another Make command called addsuffix that will add .txt to the end of all the values in YOB.
So we create the years, 1900 to 2015, we add the prefix of data/raw/yob to each of our years, so this would give us something like data/raw/yob, $(1923), but then we need to add the suffix for that text file. So we'd have data/raw/yob, $(1963.txt), and that then would be YOBTXT that would then be a dependency that we have for some of our make commands.
Back to the Makefile
- Add those three lines to the top of the Makefile
- After those lines, convert this to a rule...
# Concatenate the annual baby name data# Depends on: data/raw/yob????.txt files# code/concatenate_files.R# Producees: data/processed/all_names.csvR -e "source('code/concatenate_files.R')"And so here we have the rule to generate data/processed/all_names.csv, has data/raw/yob, the year text files. So, we can replace this with our variable from over here, which would be YOBTXT. To do this, we will have our dependency…I'm sorry, we'll have our target, all_name.csv, we will have code/concatenate_files.R, and then we will define a variable kind of like we did before which was with a dollar, parenthesis, and the name of the variable inside of it.
Back to the Makefile
- Add those three lines to the top of the Makefile
- After those lines, convert this to a rule...
# Concatenate the annual baby name data# Depends on: data/raw/yob????.txt files# code/concatenate_files.R# Producees: data/processed/all_names.csvR -e "source('code/concatenate_files.R')"
... like so ...
data/processed/all_names.csv : code/concatenate_files.R $(YOBTXT) R -e "source('code/concatenate_files.R')"And so here we have the rule to generate data/processed/all_names.csv, has data/raw/yob, the year text files. So, we can replace this with our variable from over here, which would be YOBTXT. To do this, we will have our dependency…I'm sorry, we'll have our target, all_name.csv, we will have code/concatenate_files.R, and then we will define a variable kind of like we did before which was with a dollar, parenthesis, and the name of the variable inside of it.
So the dependencies here are actually quite large. It's this file as well as all of the YOB from 1900 to 2015.txt files. And then it's going to run the command source('code/concatenate_files.R'), and this is going to look for those year of birth files. Copy and paste this in. So, we're going to copy this information into our Makefile, some I'm going to go ahead and copy the comments because I forgot to put that down.
And we'll get the rule here. And I like to have some white space between my different rules so it's easier to read, and I can save this. So I can do make family_report.html. Ah, and it's complaining about a separator again.
See, I do this a lot. Replace those spaces with a tab, quit, make, and it tells me it's up to date. So we're good to go. Other rule that we have, that you can hopefully do again, is to convert this, where we downloaded the data into a rule.
And convert this
# Depends on: website data at https://www.ssa.gov/oact/babynames/names.zip# Produces: slew of files named yob????.txt and a pdf filewget --no-check-certificate -P data/raw/ https://www.ssa.gov/oact/babynames/names.zipunzip -d data/raw/ data/raw/names.zipAnd so this is going to be kind of a weird rule, because we're generating targets which are those YOB files, but there's no dependency.
And convert this
# Depends on: website data at https://www.ssa.gov/oact/babynames/names.zip# Produces: slew of files named yob????.txt and a pdf filewget --no-check-certificate -P data/raw/ https://www.ssa.gov/oact/babynames/names.zipunzip -d data/raw/ data/raw/names.zipto this...
# Depends on: website data at https://www.ssa.gov/oact/babynames/names.zip# Produces: slew of files named yob????.txt and a pdf file$(YOBTXT) : wget --no-check-certificate -P data/raw/ https://www.ssa.gov/oact/babynames/names.zip unzip -d data/raw/ data/raw/names.zipAnd so this is going to be kind of a weird rule, because we're generating targets which are those YOB files, but there's no dependency.
This is one of the downsides of Make, is that there's no way in Make to say, hey, go look at this file up on the Social Security Administration website and see if it's been updated. And if this has been updated, then regenerate, redownload the files, regenerate the report.
So you have to be kind of…kind of to do some things yourself. So this is the rule that we'll get, we'll have YOBTXT as the target, no dependencies, but then these are the commands. So here instead of having one line of a command, we instead have two lines of a command. So I'm going to copy this into my Makefile. And there we go.
So if one of my YOBTXT files is deleted, it's going to say, "Well…" So say I delete yob 1976.txt, it's going to say, "Well, I need that here."
So it's going to come back to this rule, and it's going to then download all this information to reobtain that text file in the process as you can perhaps imagine because all those files are contained within names.zip, it's going to get all those files as well. So, that's not a big deal, but not that big, this is the way the data are stored online for us to use.
So, we'll save, quit. I got to remove those spaces. Save and quit. And then do make family_report.html, and family_report is up to date.
Sweet.
Try this...
$ make family_report.htmlmake: `family_report.html' is up to date.Sweet.
Try this...
$ make family_report.htmlmake: `family_report.html' is up to date.
What do you think will happen if we try this?
$ rm data/processed/alive_2016_annual.csv$ make family_report.htmlSo in my Schloss family report, it says at the bottom here, "if you want to use other names, you will need to comment the outlines 23 and 24 from concatenate_files.R. This will cause the scripts to run a bit slower."
So, because it would normally generate these types of data for everybody, I limited it to just the names of my kids. But what I'd like to do is add Simon to this and see what that does to the processing of our data. And so if I go into nano code/concatenate_files.R, and I look in here, so my kid's name, so in here as a vector, I'm going to add Simon.
And so now, I've changed this file, concatenate_files.R. So I quit out and I go into Make, I see that I've changed concatenate_files.R. So this is going to cause this rule to run, so data/processed/all_names.csv. And so if we scroll down we might say, "Well, where else are data/processed/all_name.csv used?"
So here it is. So it'll run this rule then, and then it will also run this rule. So if we save and back out, and we do make family_report.html, run that, you can see it's rerunning concatenate_files.R. It's then rerunning the report. And if I go back into FileZilla and reopen this, I don't see Simon in here, but that's because I never modified my report.
So, I need to then go into and modify now family_report.Rmd. And I will generate a time plot. He's a male, and data. And I will want to then also add Simon down here.
He's a male. And I need to update everybody's years a little bit. So we'll do this. Joe's actually 13. John will be 10 next week, so I'll put 10. And let's see. He's six, four, three...four.
Martha is 2, and Simon is, let's call him 0.5. And we'll quit out of this, and we'll then make family_report.html.
And we have Simon. Oh my gosh, is Simon really becoming that much more popular?
I'm shocked. So that's pretty cool, Simon is still kind of a rare name. And you can then see the data here for Simon has been added to my plot. And this is, again, cool. Like, this was what I thought about when I read the FiveThirtyEight article, was, "Where do my kids fit in here? Where do I fit in here?Where does my wife fit in here?"
And so we've been able to first replicate their analysis, and then as if you will riff on it, by converting our driver file to be a Makefile. And that's pretty cool, I didn't realize that Simon had gotten so popular as a name.
Common errors
$ make family_report.htmlMakefile:23: *** missing separator. Stop.You used spaces instead of a tab to inset your commands
So some common errors that we see frequently, we already saw this because of my copying and pasting. That if you get this error that says makefile: * missing separator. Stop, this is generally because you use spaces instead of a Tab key to inset your commands. Replace those spaces with a single tab and you should be good to go.
Common errors
$ make family_report.htmlMakefile:23: *** missing separator. Stop.You used spaces instead of a tab to inset your commands
$ make family_report.htmlmake: *** No rule to make target `family_reportRmd', needed by `family_report.html'. Stop.You mistyped the name of the dependency
So some common errors that we see frequently, we already saw this because of my copying and pasting. That if you get this error that says makefile: * missing separator. Stop, this is generally because you use spaces instead of a Tab key to inset your commands. Replace those spaces with a single tab and you should be good to go.
You might get an error that says no rule to make target family_reportRmd, see it's missing a period, needed by 'family_report.html'. Stop. So this generally means that you mistyped the name of the dependency.
Common errors
$ make family_report.htmlMakefile:23: *** missing separator. Stop.You used spaces instead of a tab to inset your commands
$ make family_report.htmlmake: *** No rule to make target `family_reportRmd', needed by `family_report.html'. Stop.You mistyped the name of the dependency
$ make family_reporthtmlmake: *** No rule to make target `family_reporthtml'. Stop.You mistyped the name of the target
So some common errors that we see frequently, we already saw this because of my copying and pasting. That if you get this error that says makefile: * missing separator. Stop, this is generally because you use spaces instead of a Tab key to inset your commands. Replace those spaces with a single tab and you should be good to go.
You might get an error that says no rule to make target family_reportRmd, see it's missing a period, needed by 'family_report.html'. Stop. So this generally means that you mistyped the name of the dependency.
You might also get something where you say make family_reporthtml, again, missing a space. And this will say, No rule to make target family_reporthtml. Stop. This means that you mistyped the name of the target in all likelihood, or that the rule doesn't exist in the Makefile.
YEARS=$(shell seq 1900 2015)YOB=$(addprefix data/raw/yob,$(YEARS))YOBTXT=$(addsuffix .txt,$(YOB))# Depends on: website data at https://www.ssa.gov/oact/babynames/names.zip# Produces: slew of files named yob????.txt and a pdf file$(YOBTXT) : wget --no-check-certificate -P data/raw/ https://www.ssa.gov/oact/babynames/names.zip unzip -d data/raw/ data/raw/names.zip# Concatenate the annual baby name data# Depends on: data/raw/yob????.txt files# code/concatenate_files.R# Producees: data/processed/all_names.csvdata/processed/all_names.csv : code/concatenate_files.R $(YOBTXT) R -e "source('code/concatenate_files.R')"# Fills in missing data from annual survivorship data# Depends on: data/raw/alive_2016_per_100k.csv# code/interpolate_mortality.R# Produces: data/processed/alive_2016_annual.csvdata/processed/alive_2016_annual.csv : data/raw/alive_2016_per_100k.csv\ code/interpolate_mortality.R R -e "source('code/interpolate_mortality.R')"So here is what our Makefile looks like at this point. Again, each chunk represents a different rule that we are using to specify the dependencies, what we're generating, and the code or the method, the command, needed to go from those dependencies to those targets. One of the things you might notice in looking at these variable names or dependency names is that there's a lot of repetition, so we have data/processed shows up frequently.
Continued...
# Generate counts of total and living people with each name# Depends on: data/processed/alive_2016_annual.csv# data/processed/all_names.csv# code/get_total_and_living_name_counts.R# Produces: data/processed/total_and_living_name_counts.csvdata/processed/total_and_living_name_counts.csv : code/get_total_and_living_name_counts.R\ data/processed/alive_2016_annual.csv\ data/processed/all_names.csv R -e "source('code/get_total_and_living_name_counts.R')"# Renders an Rmarkdown file that creates various plots and# provides an entertaining color commentary# Depends on: data/processed/total_and_living_name_counts.csv# code/plot_functions.R# Produces: family_report.htmlfamily_report.html : family_report.Rmd\ code/plot_functions.R\ data/processed/total_and_living_name_counts.csv R -e "library(rmarkdown); render('family_report.Rmd')"We can attempt to make the code DRY with variables
RAW = data/rawPROCESSED = data/processed
We can call these by using $(RAW) or $(PROCESSED)
YOB=$(addprefix data/raw/yob,$(YEARS))becomes...
YOB=$(addprefix $(RAW)/yob,$(YEARS))
Where else can you replace data/raw and data/processed?
And so what we might think about doing is it'd be nice to make this a little bit more dry, because if I instead of putting it in data/processed maybe I want to put it in data/R to denote that it's output from R or something else, then I'd have to update all these all the way through.
And so, while that might be a bit of a silly example, there are ways to create and use variables as we've already seen with the years in YOBTXT variables. And so we can make our code a bit more dry by defining two variables, raw and processed. So we might say that RAW is data raw, PROCESSED is data processed.
And then we can use these much like we already did down here with yob, where we insert years with a dollar sign parenthesis for years. So we could replace data/raw/yob with $(RAW)/yob. Because maybe we'll move where data/raw is, maybe it will become data_raw instead of having a sub directory within data of raw.
So if we look at our Makefile, we can create a raw variable RAW equals data/raw. PROCESSED equals data/processed. And so down here where we have data/raw, we could do $(RAW), and we could do RAW.
And up here. So we could go all the way through, and we could update our code to use these variable names instead of the paths.
And so up here we could define our paths, and so if we need to reorganize our project we could use that. And again, we've already seen this a bit with the years in the YOBTXT.
Automatic Variables
$@: the target$^: all dependencies for current rule$<: first dependency$*: pattern match
Can you now rewrite this?
data/processed/all_names.csv : code/concatenate_files.R $(YOBTXT) R -e "source('code/concatenate_files.R')"There are some several built-in variables within Make that we can use, and so these get a bit confusing, and I admit that I usually have to look these up, or I need to have a little cheat sheet that I keep by my computer. And so if we have a star, ampersand that denotes the target, if we have dollar sign caret, that's all the dependencies for the current rule, so the target, this could be represented as a dollar sign ampersand, or at signed I guess, with all of the dependencies in the current rule would be dollar sign caret, the first dependency.
Automatic Variables
$@: the target$^: all dependencies for current rule$<: first dependency$*: pattern match
Can you now rewrite this?
data/processed/all_names.csv : code/concatenate_files.R $(YOBTXT) R -e "source('code/concatenate_files.R')"data/processed/all_names.csv : code/concatenate_files.R $(YOBTXT) R -e "source('$<')"There are some several built-in variables within Make that we can use, and so these get a bit confusing, and I admit that I usually have to look these up, or I need to have a little cheat sheet that I keep by my computer. And so if we have a star, ampersand that denotes the target, if we have dollar sign caret, that's all the dependencies for the current rule, so the target, this could be represented as a dollar sign ampersand, or at signed I guess, with all of the dependencies in the current rule would be dollar sign caret, the first dependency.
So in this case code/concatenate_files.R would be the first dependency. And then there's something, a dollar sign star for pattern match. So how would we write this instead? So instead of having code/concatenate_files.R, we could use dollar sign less than instead of code/concatenate_files.R.
So you see we've simplifed the code, because, again, the fear is that we might change the name of the dependency and then it would come into here. It makes it a little bit tidier, at the same time, I don't try to get too carried away with this because it does obstruct the readability of my code. Because, again, like I told you, I frequently forget what these dollar sign variables are.
But if you're looking at other people's Makefiles, you'll frequently see these automatic variables, and this is the jargon that's used. These are called automatic variables, because they're built-in to Make.
Pattern matching
print-% : @echo "$*= $($*)"
The % matches a pattern. This is a weird example
Similarly, we can also use a dollar sign star for pattern matching, and the percent sign matches a pattern. And so this is a bit of a weird example, so if you type make print-, and then say YOB, that percent will match the YOB.
And then echo is a command that will use the dollar sign star to say echo equals, and then the value of the echo variable, because it'll put echo here to replace the dollar sign star, and then it will come out and we'll say, "Okay, this is a variable, let's print out the variable."
Pattern matching
print-% : @echo "$*= $($*)"
The % matches a pattern. This is a weird example
$ make print-YOBTXTYOBTXT= data/raw/yob1900.txt data/raw/yob1901.txt data/raw/yob1902.txtdata/raw/yob1903.txt data/raw/yob1904.txt data/raw/yob1905.txt data/raw/yob1906.txtdata/raw/yob1907.txt data/raw/yob1908.txt data/raw/yob1909.txt data/raw/yob1910.txtdata/raw/yob1911.txt data/raw/yob1912.txt...
Having this rule in my Makefile is helpful when I'm doing complex things like build YOBTXT
Similarly, we can also use a dollar sign star for pattern matching, and the percent sign matches a pattern. And so this is a bit of a weird example, so if you type make print-, and then say YOB, that percent will match the YOB.
And then echo is a command that will use the dollar sign star to say echo equals, and then the value of the echo variable, because it'll put echo here to replace the dollar sign star, and then it will come out and we'll say, "Okay, this is a variable, let's print out the variable."
And so if we type make print -YOBTXT, we see all of our YOBTXT files. So again, if we do make, I need to add this to my Makefile, so nano Makefile. I generally put a command like this or a rule like this at the top because it's kind of a utility function, and I like to use this as a resource to help me as I'm building more complicated variables like this YOBTXT.
So, print - : tab in, @echo "$ = $($)." So if I do make print-YOB, I get these data/raw/yob1900 all the way up to 2015.
And so again, I can then say, "Okay, that works." Now let's add the text suffix, and now I see all my paths. So, I like using this print rule to build more complex things like this YOBTXT variable. Sometimes if it says it can't find a certain variable, I will use this to see what it's spitting out.
And so sometimes it might be blank, and that tells me that I've got a problem in how I'm building my pattern. But it is a bit of a more advanced move that you might not want to worry about right yet.
PHONY Rules
.PHONY: cleanclean : rm -f $(RAW)/*txt rm -f $(RAW)/*pdf rm -f $(RAW)/*zip rm -f data/processed/*.csv rm -f family_report.html- There's no target named "clean" - this is a phony rule
- What happens if you run
make clean?
Another type of rule that we can make is called a phony rule. And so a phony rule is a rule that, it doesn't really have a target. And so here we're saying .PHONY to define it as a phony rule, colon, clean.
So we'll have a rule called clean that has no dependencies, it doesn't actually produce anything, but it will delete a whole bunch of files, it'll delete all the files you've created. And so I'm going to go ahead and copy and paste this into my Makefile. I like to put this rule at the very bottom, and this will burn down the project.
So if I save that, and if I look in my directory right now, I see that I've got my family_report.html, if I look at ls/data/raw, I've got a bunch of files there. If I do make clean, ah, it's complaining. I forgot to remove my spaces.
If nothing else, me for getting this all these times will remind you to use a tab instead of spaces, make clean. So we see that family_report.html is removed. If we use ls data/raw, those files are all removed.
So we've burned everything to the ground, and what I love about Make is now the ability to go make family_report.html, fire that off and it works. Great. And we can again look in FileZilla, see what it looks like. And we've got the updated data, and it all works, and it looks beautiful.
Other commands
make -n target: What commands will be run to build the target?make -d target: What dependencies need to be fulfilled to build the target?make –j N target: Use N processors to build targetmake: Build first rule in Makefilemake –f my_makefile: Usemy_makefilein place of Makefile
So there's some other commands that we can use with Make. We can use make –n in the name of the target, this will tell us what commands will be run to build the target. make –d in the name of the target will tell us what dependencies need to be fulfilled to build the target. make –j will allow you to use multiple processors where you define a target, and then it puts your different dependencies on different processors.
Typing make alone at the command line will build the first rule in a Makefile. So I like to specify the actual target I want to run, and if Make knows to look for capital M, Makefile, but if for some reason I name my Makefile something else I can do make –f my_makefile. And this will then use my_makefile in place of your name to Makefile.
General "design" thoughts
- Scripts are better than command line calls in the Makefile. The Makefile isn't a dependency!
$(YOBTXT) : wget -nc --no-check-certificate -P $(RAW) https://www.ssa.gov/oact/babynames/names.zip unzip -u -d $(RAW) $(RAW)/names.zipSo, some general design thought so to speak, I think using scripts are better than command line calls in the Makefile, so the Makefile isn't a dependency itself. So say the SSA website changed, well, I could change it here, but that doesn't mean it's going to regenerate YOBTXT, those files, because there's no dependency here. But if I had put this all into a script and then made the script a dependency, then if I updated the code, then it would regenerate YOBTXT because the dependencies are newer than the target.
General "design" thoughts
- Scripts are better than command line calls in the Makefile. The Makefile isn't a dependency!
$(YOBTXT) : wget -nc --no-check-certificate -P $(RAW) https://www.ssa.gov/oact/babynames/names.zip unzip -u -d $(RAW) $(RAW)/names.zip- Make scripts specific to one function. This minimize the scope of the dependency tree. If you change one script, you will affect far fewer downstreame files
So, some general design thought so to speak, I think using scripts are better than command line calls in the Makefile, so the Makefile isn't a dependency itself. So say the SSA website changed, well, I could change it here, but that doesn't mean it's going to regenerate YOBTXT, those files, because there's no dependency here. But if I had put this all into a script and then made the script a dependency, then if I updated the code, then it would regenerate YOBTXT because the dependencies are newer than the target.
If you make a script try to make it specific to one function, this will minimize the scope of the dependency tree. So if you change one script, you'll affect fewer downstream files. So for example, if you're doing an analysis and you put all of your R code into one file to generate say 5 or 10 different plots, and to run all different statistical analysis, if you update that file then everywhere that you call that file is going to run, but if you make a separate file for each figure in your paper then updating one figure isn't going to cause you to regenerate all the other figures. And so again, I like this because it keeps things more compartmentalized and easier to work with.
Some other resources in case you're interested include Software Carpentry, they've got a lot of great resources, and one of the tutorials they have that I really like is on Make, it's basically where I learned to use Make. GNU make is the reference for using Make. And then Stack Overflow is often times pretty useful too. Make can be confusing, it can be frustrating, but again, all the stuff with the variables, the automatic variables or the variables you name, those are all extra, they're not that critical. If you long-hand write out the names of your dependencies and your targets, you'll be in great shape.
It's kind of I find that when I try to go to the next layer and think about things like variables that I cause problems for myself. All right.
Let's turn back to Kozich
- In the project directory there is already a
Makefile- Can you identify the variables?
- Can you see the rules for the bash commands you put into
analysis_driver.bash? - Does anything need to be edited for this project?
- What is missing?
So we're not here to study babies and their names, we want to turn back to Kozich analysis. So, in the project directory there's already a file there called Makefile, and so what I want you to do is see if you can identify the variables.
Let's turn back to Kozich
In the project directory there is already a
Makefile- Can you identify the variables?
- Can you see the rules for the bash commands you put into
analysis_driver.bash? - Does anything need to be edited for this project?
- What is missing?
Either adapt this Makefile for the Kozich project based on what you learned from the 538 name tutorial or delete the file and start over
- Rememeber that
make -n <TARGET>allows you to do a dry run
So we're not here to study babies and their names, we want to turn back to Kozich analysis. So, in the project directory there's already a file there called Makefile, and so what I want you to do is see if you can identify the variables.
Can you see the rules for the bash commands you put in the analysis driver bash file? There are some things already in there, because, again, this came out of the template for a new project, and a lot of the things are pretty common. And so what type of things need to be edited for this specific project and what is missing? And so I would encourage you to either adapt this file for the Kozich project based on what you learned from the FiveThirtyEight name tutorial, or delete the file and start over.
And remember that writing make –n allows you to do a dry run and see what happens. And so as you go through the Kozich file, recall that when you're getting the fastq.gz files don't delete the tar file, that that perhaps could be your target for that download, instead put it in data/raw and make it that target.
Hints...
- When getting the fastq.gz files, don't delete the tar file
- Instead, put it in
data/rawand make it a target - Use the tar file as a dependency in the next step
- Why isn't this ideal?
We can use the tar file as a dependency in the next step. And so you might ask, "Why isn't this ideal?" And that's something for you to think about. And so as you go along, I want you to think about make –n on various targets along the ways, and what all needs to be done. And then finally, if you remove your silva.seed align and you write make –n write.paper, what is going to happen?
What happens if you do...
make -n data/references/silva.seed.alignmake -n results/figures/nmds_figure.pngmake -n write.papermake -n HMP_MOCK.v4.fastarm data/references/silva.seed.alignmake -n write.paperNano Makefile. And so at the top here you'll see some variables, refs, figs, tables, proc for processed, final for submission directory, the helper function for printing. There's also three things in here that I'm going to go ahead and remove, I'm going to comment out and I encourage you to do the same, because it seemed like a good idea at the time.
But they just seem to cause problems, so I'm going to delete those, out of sight, out of mind. And so we look through here, we see downloading of our reference files. Here we're using the RDP trainset. Up here we've got our silva reference alignment, how to run the data through Mothur. We have our basic stem, and the filenames get quite long, so we can use basic stem to replace that if we want.
We also then have get_good_seqs.batch, along with the dependencies. And again, here are our get_shared_otus.batch and notes what you need to update, which we've already updated when we were doing our analysis of driver batch file.
And this then is getting the error, and there's also a part here for figures and then also for building our manuscript. So, excellent. So we need to add a few things. So we need to add a few things, so we need to get the data into here. Some I'm going to come back, I'm going to exit out. And I'm going to do a nano analysis_driver.bash, and we want to get this chunk here for our stability, WMetaG.tar file. nano Make.
I forgot to save when I quit before, so I'll remove those files. And if we come down to the top of part two, I'll put it in here. And, the suggestion was to make data/raw/StabilityWMetaG.tar our target.
It won't have any dependencies, that. And instead of removing StabilityWMetaG.tar, we're going to move this to data/raw. So what's going to happen is it's going to download StabilityMetaG.tar to our home directory or project directory.
It's going to untar it and throw the data into data/raw. It's going to then move our tar file into data/raw. And so then if we run it again it will see that that is there. So if I do…I'm going to copy that, because I always forget names within like three seconds.
So the other thing that we want to bring in is our figure, our ordination. So if we nano analysis_driver bash. So down here, Constructor NMDS png file. So we'll copy that into out Makefile. So that was down here in part three.
And formatting gets kind of wonky. So that's good. All right, and so the output, we need to know the target.
And the dependency. And then this is the code we're going to run. So one dependency is going to code/plot_nmds.R. The other is going to be this horrifically long name.
And we saw up above that we can replace everything through pre-cluster with our base renewable name. And so I need to double-check what that was because like I said I'm forgetting. And so if I move up, basic stem.
We'll do (BASIC_STEM), so that's that. And we can then also replace this horribly long name. That's the other advantage of the variables, that if we've got these really long filenames we can simplify them a bit with using stems of variable names.
And so, get that working. So our dependency was this, so we can take that out. The other thing we could do is we could reverse the order of these dependencies so that instead of having to write out the long name we could use the automatic variable.
I'm not totally sold on the automatic variables as I mentioned because I frequently forget what they are. I'm also forgetting what the name of our target is, so I'm going to ls. ls results, figures/nmds_figure.png. So, it's results/figures/nmds_figure.png.
Go back into my Makefile, and so it is results/figures/nmds_figure.png. Down in the manuscript section, we need to add the dependencies, so if you recall our RMD file inserts a copy of our ordination figure, and I'm going to copy and paste the name of that file just because I always forget these things.
So, results/figures/nmds_figure.png. And also we brought in a data file to our code chunk, so let's go back to our manuscript RMD submission directory, and let's grab this shared file, because this is a dependency for making the manuscript.
So we fly back down, and pop that in. And again, we want the backslash there, and recall that we can replace all this jazz with the stub, the BASIC_STEM. So these are some extra dependencies for rendering our manuscript.
Also down here in write.paper, where we want to make sure that we have all the things we need in order to submit the manuscript, we're going to go ahead and delete these files, and we're going to add in results/figures…see I forgot, /nmds_figure.png.
And so we should be good. So back here if we do make –n write.paper, make: Circular submission//manuscript.Rmd so submission//manuscript.Rmd dependency dropped.
Again, that's not a big deal, if we look at our Makefile, again, go to the end, so to run write.paper we need all these RMD files, and so it uses that percent sign to match any of the extentions. So, it's trying to build manuscript.Rmd which it can't do because it's got itself as a dependency.
That's a circular rule, and so that's not allowed. And so it's not going to build that, which is fine by me, but it's going to build a Markdown file, a tex file, and our manuscript.pdf file. And so again, we see the commands it's going to run, which are the commands for write.paper.
So we do make write.paper. It's running through, it's building all of this, and everything is good. So if we do make –n write.paper, we see there's nothing to be done. It's also complaining about that dependency. And if we do lslth submission, we now see that our pdf, our tex, our md files are all newer than our R Markdown, bib, tex, and csl files.
So if our Rmd was newer, it would rerun and regenerate those. So, there's a couple things that we could do to make this better, we could add a rule to download Mothur, we could make Mothur a dependency on all of these, or we could, like I said, we could change the reference database we used and other things.
Final touches
- Write a PHONY target:
clean - Commit!
- Run
make clean; make - For all the marbles...
cd ..rm -rf Kozich_ReAnalysis_AEM_2013git clone ...cd Kozich_ReAnalysis_AEM_2013make write.paperWhat I'm going to do to prove to ourselves that this works is I'm going to go look at ls data/raw, and we see all those gz files. I don't have a phony, I don't have a clean rule. You could you could make a clean rule. I'm going to rm data/raw/gz data/raw.tar. ls data/raw, and we see that that's pretty empty.
So if we went ahead and we ran Make, then everything make write.paper, it should work.
So, before we do that, let's go ahead, and… You know what? I'm going to go ahead and add that rule, that Mothur rule to my…I'm going to go ahead and add that Mothur rule from my analysis_driver.bash, to my Makefile. And so that's this first line here.
And so, exit out nano Makefile. And I'll put that at the very top here. And again, the formatting gets kind of funky. And that dependency is going to be code… I'm sorry, the target is going to be code/mother, there's no dependency, we need to tap these all over.
And again, what we could do is we could make, not there, but here we could make code/mother a dependency. And we could do that for all of these as well.
So what do we do now? Well, let's commit this, we've done a few things to modify our project. We do git status, we've modified our Makefile, we've modified md and pdf. I go ahead and git add make, and submission/manuscirpit.md, and submission/manuscript.pdf, git commit.
We're going to transfer... I'm going to also git rm analysis_driver.bash, and git commit is "Transfer from analysis driver script to Makefile."
So that's commited. git status, git push, enter our credentials. Excellent.
So, we're going to burn this thing down. So I'm going to do rm-rf Kozich. Oh, that was scary. So now we want to prove to ourselves that we can clone it, so we github/pschloss/ Kozich_ReAnalysis_AEM_2013, I'm going to clone this, copy that. git clone.
Cd into Kozich, ls, ls submission. We've got our manuscript files, but if we ls data/raw, there's nothing there. So what we're going to do, and this awesome, is we're going to make write.paper.
And so before you hit Enter, make sure you're in tmux, because it's going to take a little while to run. We hit Enter. And it complains, No rule to make target 'data/mother/stability…' Ah, because we forgot a rule. But you know what?
We deleted it. So we deleted our analysis_driver.bash file. So what we want to do is let's go back and use our history, I perhaps got a bit aggressive in deleting that. So I'll go to my previous commits, where we transferred from analysis_driver script to Makefile. And I see that I deleted my analysis_driver.bash, and somewhere in here I get_nmds_data.batch.
We can copy those lines. I'm not going to worry about get_sobs_data.batch, because we didn't use any of that data in our Makefile. So I do nano Makefile, and come down. I guess I should have done the make test before I did everything else, but that's okay.
It's better if I put this with my figure and table generation. And, this is the rule that the target is this long beast. And the dependency for that is our shared file, which is up here, right here.
And we can bring that down. This becomes our dependency, and then that is our command. Quit, and again, if we try make write.paper, it's firing away.
So to get out of tmux we can do Ctrl + B + D, and we're back to our Make tutorial directory. But whatever if we, cd ~/Kozich, we can sit tight, and knowing that we can go back in a tmux a and watch it at any time. And sure enough we see another error, mothur: not found, and that's because our Makefile doesn't know that Mothur is in code Mothur.
So this would be a great variable to make. So MOTHUR, we will do code/mothur/mothur. So, I think we gave it the wrong directions all the way around, it's supposed to be code/mothur/mothur. And so if we put the executables in the Mothur directory, and we can replace this code/mothur with $(MOTHUR).
And I'm going to copy this to make it easier to copy and paste throughout. Then I'll go ahead and make this a dependency here. This all looks good.
I'm just looking for cases of where we're calling the mothur executable, and there is one. And again, we can add this as a dependency.
All right. And I think there's one more.
So we can do a search if you…Ctrl + W for search, we can type MOTHUR, and we can look for cases of the lower case mothur, where it's not supposed to be.
So I'm going to replace this with our variable name. We're looking good like we got all the cases. Yeah, I think we've got it. All right, so save that, and quit.
No we'll do make write.paper, and we'll just watch it, and it'll wrip. So it looks like we got an error here, where it doesn't like my use of wildcards in tar, so I'm going to go ahead and enable these wildcards in my tar.
And then I'll make…and go look for a trainset, so we can wget trainset. And then save this and make write.paper. And… Is that what it told me to do? Use, we'd have to put the wildcards before the xvzf. I'm having a hard time seeing what's going on, so if I make that. All right. I'm not sure what's going on, I'm going to delete trainset.
Now let's try it again. All right. So I'm just going to remove this wildcard thing, I was getting kind of cute.
So we delete that, and try make write.paper again. This is real life, folks. So, basically what happened is that before I had this tar x, gz where I'd use a wildcard to only pull up the files I wanted.
And so, for some reason, the version that's on Amazon of TAR doesn't like that, and so I went in and I removed that star at the end that you saw. But now it's complaining because I have a rmdir to remove this directory, but it's not empty.
So we need to modify that, instead of undo I'm going to do rmdir –rf. So we can do rm –rf, that, make. Oh, and so I type make, and the first rule like we saw ran, but that's not what I want, I want make write.paper.
data/mothur. Ah, there's a bunch of log files in there. Let's get to this. So, unable to find any gz files in your directory.
Is that because it hasn't downloaded them? I think that's what it is. So if we do ls data/raw, we don't have any gz files.
So we don't have our dependencies straight, and so we need to tell it what to do. So basically, we set this as a rule, as a target, but we never use that as a dependency, so we need to add that as a dependency. So, we will come down here, and we will add that as a dependency, save that.
We will make write.paper, and now it's going to download the data. So just because we make a rule doesn't mean it gets called, so we have to use the target for that rule as a dependency elsewhere or we have to make it explicitly for it to get used. And so this is going to take a while to go, but I'm confident that it's going to work this time around.
Well, after a few hiccups that finally worked through and we got to the end, and we can also do ls –lth. And we see that, in submission that our files have been generated and opened, I can go into FileZilla, and I have to go back up to a higher level, to my Kozich_ReAnalysis, submission, manuscript.pdf.
And similar to what we had in the previous tutorial, we see that we've got our paragraph in here, our plot is embedded, and it all looks great. So there were a few hiccups along the way of course in getting that to work, but I think that also underscores the value of doing the make clean and then make write.paper again to make sure that our ducks in a row and everything works and that it's reproducible for us.
If you're seeing this tutorial or if you started the tutorial series after April 25th, you probably have a set of code for the new template that won't have the problem, many of the problems that I ran into. I've gone back and I've updated the code that comes in the template so that it works a little bit better here on AWS than what we experienced.
Regardless, I still think that it's really valuable for people to see how we troubleshoot what's going on with our code, and why we get things that don't work, and ultimately getting it to work for us is making it more reproducible for others. If it's reproducible for us it's going to be reproducible for others as well.
So let's go ahead and do git status. And we've got our updated files. Something we might do is we could do git diff –word submission/manuscript.md. And we see that there is a small…the only difference between the two files, between this run and the previous run is about 17 sequences on average, and this is probably coming through because of the randomness in our chimera checking, and classification a few of the other things.
So we will go ahead, and again, we did the git status. We'll do git add Makefile. Something weird here happening with the data/references/README, I think it's written over what we had before, we didn't change anything there. So I'm going to check that out and revert to the old version.
So we do git checkout--the name of that file, git status. And then we can do git add Makefile results/figures/nmds_figure.png submission/manuscript.md submission/manuscript.pdf submission/manuscript.tex. Those have all been staged, git commit.
And we will say…what should we call this? What did we do? We modified the Makefile to get it to work, workflow. Great, and now we can push it.
Enter our credentials. And excellent. So again, what we've done is we've demonstrated to a pretty good degree that we can burn down our directory, we can clone it from GitHub, we can then write make write.paper, and voilà, we can recreate it. So I'm going to go ahead and exit out of tmux, exit out of Amazon.
One last thing...
- The same samples were sequenced twice and were posted at the same website that you obtained your data from
- What would you change in the Makefile to re-run the analysis?
- How could you test the effect of the change to the overall analysis?
- Give it a shot and see how far you get
- Make sure to commit your repository before you change the raw data
- Modify your Makefile and remove its target
- Run
make write.paper
- Run
git diff-word submission/manuscript.md - What changed?
And quit that, and then we're going to go into Amazon, and we're going to stop our instance. If you have the time and the interest, I'd encourage you to go back and do a small experiment, and that would be to go back into your Makefile and change what dataset you're processing.
If you recall back from when we pulled the data off of the Mothur website to get that tar file, there were two options, one was [inaudible] data that was sequenced with shotgun metagenomics data. And the other was without shotgun metagenomics data. And so go ahead and modify your Makefile and trigger it to generate a new, to rerun that rule that you modify.
And then rerun it and see what happens, of course that's going to take an hour or so to run, and at the end you can run git diff –word submission/manuscript.md and see what changed in those sentences.
Parting thought...
make clean; makeis a philosophy to work by- Probably more important than actually using
make - You won't be able to put all of your data into git/GitHub
- Instead assume that someone will get your Makefile and the skeleton of your project
- Probably more important than actually using
Parting thought...
make clean; makeis a philosophy to work by- Probably more important than actually using
make - You won't be able to put all of your data into git/GitHub
- Instead assume that someone will get your Makefile and the skeleton of your project
- Probably more important than actually using
- Don't worry about variables and fancy
makestuff: get it to work and then refactor
As a parting thought, I'd really like you to think about the philosophy that we just exhibited, and had a little struggle with but got it to work, of make clean make, that that process of again burning down our directory, of deleting it, pulling it down, or deleting all the processed files we made, and then rerunning it back through to make sure that it's reproducible is really a great philosophy to work by.
If you approach a project knowing that you are going to intentionally destroy your project and rebuild it, you will change how you approach the project, how you approach your documentation, how you approach your scripting and your coding and your automation, and that's going to be really invaluable.
Whether or not you use Make, that philosophy is really important. We haven't done anything new here with Make, the main difference of course is that we use Make because it keeps a timestamp, it keeps track of dependencies, we could have just rerun the whole script like we did previously. But you know, we had make and so that was a little bit more sophisticated.
And as our projects get bigger and more involved, we perhaps don't really want to burn down the whole project and start over again. Also, don't worry about all the variables and fancy make stuff, get it to work and then refactor from there. Like I mentioned, like some of the variables, like the automatic variables, they're not easy to read, and if it's not easy to read, it's not going to be ultimately reproducible or transparent to other people that come along.
Exercise
The analysis and writing of the original Kozich paper was done using a more "traditional" method. We went back and rewrote the paper to make it reproducible using the methods outlined in the previous modules of this series.
Look at our GitHub repository and see how the full project was structured in terms of documentation and organization, how we used automation with Make, and how versioning was used to track our progress in the rewrite.
- How does the rewritten manuscript compare to the published version?
- What could be improved in the documentation of the rewritten manuscript?
- What's the next step towards reproducibility that you feel comfortable taking?
So, I'd also like you to think about this exercise, that the analysis and writing of the original Kozich paper was done using a more traditional method, where we had a bunch of scripts running things, we didn't really publish the scripts, we tried to be transparent in our writing of the manuscript, but as we saw from the number of reads that got through, we weren't sure if that was the number of raw reads or processed reads, millions of sequences.
But I did then go back and rewrote the paper to make it reproducible using the methods we've been talking about in this tutorial series. So if you look at our GitHub repository and see the full project was structured in terms of documentation organization, think about how we used automation with make and how versioning was used to track our progress in the rewrite.
So, how does the rewritten manuscript compare to the published version? And what could be improved with the documentation of the rewritten manuscript? And then what would be the next steps towards reproducibility that you feel comfortable taking? You might start small with Make. In general when I write a Makefile, I'm not repurposing an analysis driver script.
I start with one rule, and then I add another when I'm ready to the next step, and I keep adding rules as I'm doing subsequent steps. So here what we've done is a little bit artificial because we've refactored previous code to generate what we've done, both with the baby name example as well as with the Kozich analysis. Well, think about instead starting with a blank file, we are slowly adding rules to build up your analysis.
That also is another way of approaching it that might make things a little bit easier to understand what's going on. Well, that was a lot of material, but it was really important. Next to version control and literate programming, Make is one of the three most important tools I have in my reproducible research tool box. The ability to write make clean, make write.paper is a great feeling, maybe just a little bit better than writing git in it.
Last year I wrote a paper in mBio about the use of preprints among microbiologists, it was a commentary that had some data in it and a few plots. This manuscript was written using the same methods I'm outlining in this tutorial series. Over the past two years as you may know, preprints have really grown in popularity. Well, during the time that the manuscript was in review and I was making my edits, a lot had changed in terms of the number of preprints that were being posted.
Before my final submission, I was able to use make write.paper to refresh the project and have bleeding-edge data in the paper. I did this all with minimal interaction of the code, and it was pretty awesome and underscored the value of using tools that facilitate the reproducibility of our analyses. In a couple of months, I'm going to be giving a talk at the micro meeting on preprints among microbiologists, and I look forward to refreshing the project again using make write.paper to update my figures and update the statistics that I reported about the use of preprints among microbiologists.
In the next tutorials we'll be discussing the tools that we've already been using to help foster collaboration. In the next tutorial, we'll use a process called git flow to collaborate with our subs. In the subsequent tutorial we'll discuss the use of poll requests of other resources hosted on GitHub to foster collaboration with others. I look forward to talking to you then.