Issues in Reproducible Research
http://www.riffomonas.org/reproducible_research/reproducibility/ Press 'h' to open the help menu for interacting with the slides
Welcome back to the Riffomonas Reproducible Research Tutorial Series. If you missed the introduction tutorial, please go back and check it out to learn the background behind the series, and to familiarize yourself with our goals and where we're going to be going.
As we go through these tutorials there will be several discussion items that you should discuss over lunch, at your lab meeting, or individually with your research group and with your PI. For many of these questions, there are no "right answers." The concepts we'll cover in these tutorials are difficult, that's part of the reason why there's a problem with reproducibility in science. Hopefully you can have a good discussion on these questions within your research group.
There will also be a number of exercises for you to work on where there is a correct answer. For these, remember that you can always hit the P key on your keyboard to pull up the presenter notes, which is where you'll find the answer to these questions. Also, when these come up you should pause the video to give yourself a few moments to think about the answer and to explore the materials further.
In future tutorials there will be hands on coding exercises that we'll work on together. Of course, you'll want to do each of these activities to build your own background and expertise in handling issues that surround reproducibility. At the end of the tutorial series, there will be an opportunity for you to document your progress and receive a virtual badge and certification that you can include to document your participation in this tutorial.
As I mentioned at the end of the last tutorial, today's tutorial is appropriate for those doing the analysis as well as those whose job is to primarily supervise scientists in their analysis. So let's go into today's material, which you can find in the Issues in Reproducible Research tutorial at riffomonas.org. Wonderful. Let's go ahead and pull up the slides for this session's tutorial.
And remember, we can get to the slides by going to riffomonas.org, clicking on Training modules, followed by Reproducible Research. This pulls up the Reproducible Research Tutorial Series page. Or along the left-side in the middle of the screen are the listing of the various tutorials.
Learning goals
- Summarize the origins of the "reproducibility crisis"
- Differentiate between reproducibility and replicability
- Identify pressure points in making your work reproducible
- Appraise ongoing and published research products for hallmarks of reproducible research
For today's tutorial, what we're going to focus on are several learning goals. The first is to discuss the origins of what's being called the reproducibility crisis. We want to differentiate between reproducibility and replicability.
We're going to identify the pressure points in making our work reproducible. And then we're going to appraise ongoing and published research products for hallmarks of reproducible research.
Before we go any further...
- You need to read...
- They're short and important takes on where we are in science, in general, and microbiology, in particular.
But before we go any further, I want to confirm that you've already taken a look at the Collins & Tabak, editorial in Nature, the Casadevall et al, editorial in mBio, and the editorial by Ravel & Wommack that was published in Microbiome.
They're short, but they're important takes on where we are in science in general and in microbiology in particular. So, perhaps you're familiar with this fictitious journal, The Journalof Irreproducible Results. I know when I was working in the lab more, we frequently joked about people whose theses would wind up in this journal, that we would do great things in the lab, get really excited about it, but before we told our PI we went, "Well, let's just do it one more time," and sure enough it wouldn't work out, and that we had forgotten some control or we'd forgotten some reagent, and so the work just didn't reproduce.
And so the goal of this series of tutorials is really to avoid anyone thinking that their work belongs in The Journal of Irreproducible Results, that we want all of our work to be reproducible because we want others to be able to follow up on what we're doing, so that we can move science forward.
Definitions
- Reproducibility: "Ability to recompute data analytic results given an observed data set and knowledge of the data analysis pipeline."
- Replicability: "The chance that an independent experiment targeting the same scientific question will produce a consistent result."
Definitions
- Reproducibility: "Ability to recompute data analytic results given an observed data set and knowledge of the data analysis pipeline."
- Replicability: "The chance that an independent experiment targeting the same scientific question will produce a consistent result."
Beware! Depending on who is talking, the definitions may be flipped
And so two definitions that we're going to build from in this series of tutorials for reproducibility and replicability come from Jeff Leek and Roger Peng's editorial in the journal PNAS, published in 2015. In that, they define reproducibility as the ability to recompute data analytic results given an observed data set and knowledge of the data analysis pipeline.
Okay, so find out how you analyze the data, I have your data, can I get the same results? Replicability in contrast is the chance that an independent experiment targeting the same scientific question will produce a consistent results. And so if somebody else does the experiment and uses our same general approach, do they get the same results?
But beware, because depending on who's talking, the definitions that we're using here might be flipped. And so always be careful about how people are defining things. I find this is kind of similar to how many people talk about diversity in the microbiome literature. And I always wonder, do they actually measure diversity, or do they mean richness, or what exactly do they mean?
Kirstie Whitaker (doi: 10.6084/m9.figshare.5440621)
So the same is true if we're thinking about reproducibility and replicability. Another framework that I like to think about comes from a talk given by Kirstie Whitaker that she posted on Figshare, that builds off of what Leek and Peng talked about. And so, using the same or different code, and using the same or different data, you can think of research as being reproducible, replicable, robust, or generalizable.
I like this framework a lot, because it really helps me to think about and contextualize different attempts to validate other people's research.
And so I've built upon it a little bit, so instead of thinking about the same or different data, I think about the same or different populations or systems. So, is the work done in mice versus humans?
Is it done in different strains of mice? Is it done in different cohorts of humans? Is it done using the same or different cell lines? Along the rows we can think about the same or different methods. Do they use the 16S rRNA gene sequencing? Do they use metatranscriptomics? Do they use metabolomics?
Do they do culturing? Do they use the same or different methods? And from that then, if you use the same population or system and the same methods, then we would hope that the work would be reproducible. In contrast, if you use a different population or system, say instead of using people from Michigan in your study, you use people from Korea, is the work replicable? Similarly, if we use multiple methods to triangulate on a result, to increase the robustness of that result, we can then think about doing that in different populations or different systems to then test whether or not our result is generalizable.
Pop quiz: Reproducibility or Replicability?
- A new person joins the lab and tries to repeat a previous lab member's experiments
So as a pop quiz, what I'd like you to do is to think about different questions that I'm going to flash up here as being reproducible or replicable. And so if you want, you can hit the pause button to think about it for a minute before getting the answer from me. Okay, so a new person joins your lab and tries to repeat a previous lab member's experiments. That is an example of reproducibility.
Pop quiz: Reproducibility or Replicability?
- A new person joins the lab and tries to repeat a previous lab member's experiments
- You download data from another lab, following their methods, you try to regenerate their results
So as a pop quiz, what I'd like you to do is to think about different questions that I'm going to flash up here as being reproducible or replicable. And so if you want, you can hit the pause button to think about it for a minute before getting the answer from me. Okay, so a new person joins your lab and tries to repeat a previous lab member's experiments. That is an example of reproducibility.
You download data from another lab, follow their methods, and you then try to regenerate their results. That also is an example of reproducibility.
Pop quiz: Reproducibility or Replicability?
- A new person joins the lab and tries to repeat a previous lab member's experiments
- You download data from another lab, following their methods, you try to regenerate their results
- You rerun the mouse model that you ran in a previous paper but use a different strain
So as a pop quiz, what I'd like you to do is to think about different questions that I'm going to flash up here as being reproducible or replicable. And so if you want, you can hit the pause button to think about it for a minute before getting the answer from me. Okay, so a new person joins your lab and tries to repeat a previous lab member's experiments. That is an example of reproducibility.
You download data from another lab, follow their methods, and you then try to regenerate their results. That also is an example of reproducibility.
Next, you rerun the mouse model that you ran in a previous paper but using a different strain. This in contrast is an example of replicability.
Pop quiz: Reproducibility or Replicability?
- A new person joins the lab and tries to repeat a previous lab member's experiments
- You download data from another lab, following their methods, you try to regenerate their results
- You rerun the mouse model that you ran in a previous paper but use a different strain
- Someone performs your study using a cohort of subjects in Korea
So as a pop quiz, what I'd like you to do is to think about different questions that I'm going to flash up here as being reproducible or replicable. And so if you want, you can hit the pause button to think about it for a minute before getting the answer from me. Okay, so a new person joins your lab and tries to repeat a previous lab member's experiments. That is an example of reproducibility.
You download data from another lab, follow their methods, and you then try to regenerate their results. That also is an example of reproducibility.
Next, you rerun the mouse model that you ran in a previous paper but using a different strain. This in contrast is an example of replicability.
Someone performs your study using a cohort of subjects from Korea. That is an example of replicability or perhaps generalizability.
Pop quiz: Reproducibility or Replicability?
- A new person joins the lab and tries to repeat a previous lab member's experiments
- You download data from another lab, following their methods, you try to regenerate their results
- You rerun the mouse model that you ran in a previous paper but use a different strain
- Someone performs your study using a cohort of subjects in Korea
- A colleague asks for your raw data and any scripts you may have to repeat your earlier analysis
So as a pop quiz, what I'd like you to do is to think about different questions that I'm going to flash up here as being reproducible or replicable. And so if you want, you can hit the pause button to think about it for a minute before getting the answer from me. Okay, so a new person joins your lab and tries to repeat a previous lab member's experiments. That is an example of reproducibility.
You download data from another lab, follow their methods, and you then try to regenerate their results. That also is an example of reproducibility.
Next, you rerun the mouse model that you ran in a previous paper but using a different strain. This in contrast is an example of replicability.
Someone performs your study using a cohort of subjects from Korea. That is an example of replicability or perhaps generalizability.
Finally, a colleague asks for your raw data and any scripts that you may have to repeat your earlier analysis. This is similar to the second question here, and is an example of reproducibility. reproducibility
What are some threats to reproducibility?
So something we might think about are, what are the threats to reproducibility? What are the things that stand in the way of someone else trying to reproduce our work? And so, what I'd like you to do is go ahead and pause the video, and jot down a list of bullet points or different things that you think can limit the ability of another person to reproduce your work or limit your ability to reproduce another person's work.
So go ahead and and take as much time as you need to generate that list.
What are some threats to reproducibility?
- Data are not publicly available
- Incomplete methods section
- Operating systems
- Evolution of software/databases
- Methods rabbit hole
- Use of random number generators
- Missing details in protocols
- Availability of software
- "Custom XXXXX scripts"
- URL/e-mail rot
So something we might think about are, what are the threats to reproducibility? What are the things that stand in the way of someone else trying to reproduce our work? And so, what I'd like you to do is go ahead and pause the video, and jot down a list of bullet points or different things that you think can limit the ability of another person to reproduce your work or limit your ability to reproduce another person's work.
So go ahead and and take as much time as you need to generate that list.
Here's the list of things that I came up with. Again, this is not the perfect list, it's not exhaustive, and it's perhaps particular to what I'm thinking about when I came up with the list.
So many problems we have with data being available, method sections being incomplete, different operating systems behave differently. Sometimes different programs are available on different operating systems but not another. We know that our software and databases evolve.
Mothur, the software package I develop is on version 40. It has evolved a lot over those 40 versions, and so the results you might get from version 1 to version 40 might be a little bit different. There's also the problems with methods rabbit holes, where you read a method and they say, "Well, we used…" You read a paper and they say, "We used this method."
And so then you go to that paper describing that method, and they say, "Well, this method is a mash up of two other methods." And so, and you just keep going down and down and down the citation ladder, and as you go further and further down you start to question, "What exactly did those original researchers do?" There's the issue of random number generators, a lot of times in bioinformatics we're doing things like bootstrapping or Monte Carlo simulations to calculate P values, and so those use random number generators.
And so we might get slightly different results each time we run the analysis. Similar to the problem with short method sections and rabbit holes, is that there's frequently details missing in a protocol. The availability of software. That we know that many times people will say that the code is available upon request.
There's the problem of custom fill in the blank scripts and filling in the blank with different programming language. So if I said I...you know I did my analysis using custom Pascal scripts, but if nobody knows Pascal then how reproducible is that going to be? There's also a problem of link rot, whether a URL or email address may no longer be accessible.
So you go to contact the original researchers for the data or for the code, and you get an error message back saying "Sorry, that email address is no longer valid."
What are some threats to replicability?
Next, let's think about some threats to replicability. Again, the ability to get the same result using a different population or system.
So again, take a few minutes and jot down several ideas for things that might limit our ability to replicate another group's research.
What are some threats to replicability?
- Not a real effect
- Models that overfit the data
- Poor experimental design
- Contaminated/mistaken reagents
- Confounding variables
- Sex
- Age
- Mouse genotype
- Differences in reagents, populations, environment, storage conditions
- Sloppiness
- Selection and experimental bias
- Fraud/Scientific misconduct
Next, let's think about some threats to replicability. Again, the ability to get the same result using a different population or system.
So again, take a few minutes and jot down several ideas for things that might limit our ability to replicate another group's research.
So again, my own list, not the perfect list, not exhaustive, but list of things that I think a lot about in terms of replicability.
So first, it may not be a real effect. But the first study that found the result may have been mistaken, it may have been reproducible, but it might not have been a real effect. There might be problems with mathematical models, statistical models that we're using that over fit the data in the original study that then just can't be validated in a replication study.
There's a problem of poor experimental design where things aren't controlled for that needed to be. Problems with contaminated or mistaken reagents. You know, we think we have the cell line or the specific strain of bacteria only to find out later that we were sent the wrong strains. Problems with confounding variables, variables that we're just not controlling for because we don't think about for some reason.
There's differences in sex, we might do an experiment using male mice, someone else might do it using female mice. And we might get different results because it's due to differences in sex. And the same thing might happen with differences in age, or differences in mouse genotype. And those factors of thinking about sex, age, and genotype are actually pretty interesting, they raise up other biological questions that we might be interested in following up.
There might also be differences in reagents or populations, environments, storage conditions. There also might be the problem of sloppiness, where the original researchers may have not been thinking thoroughly about what they're doing. We might also be thinking about poor laboratory skills, contaminated reagents again, things like that.
There's also a problem of selection and experimental bias. That we see a result in the literature and we then want to test it with our own population, our own system, and what we don't know is that 20 other people have tried the same thing and have failed to replicate it as well. All right, and so that one study stands out because it was a huge result, but the fact is that it stands out because nobody published all the negative studies.
And then there's the problem of experimental bias, where we find a result and now we go looking for further proof of that result, not trying to find proof against the result. And of course there's always the concern about fraud and scientific misconduct. So I hope you might see yourself or have experiences with all of these bullet points of threats to reproducibility and replicability.
This stuff is really hard!
And the point I want to make is that this stuff is really hard, science is hard. It is difficult to do reproducible research that others can replicate. There's no getting around it, it is hard.
And so this brings us to the editorial by Arturo Casadevall and his colleagues that is questioning the quality of research in the biological sciences, and specifically the problems of reproducibility in microbiology.
And this grew out of an American Academy for Microbiology report, from their colloquium that they did discussing problems in the sciences, and thinking about reproducible and replicable research.
And so this brings us to the editorial by Arturo Casadevall and his colleagues that is questioning the quality of research in the biological sciences, and specifically the problems of reproducibility in microbiology.
And this grew out of an American Academy for Microbiology report, from their colloquium that they did discussing problems in the sciences, and thinking about reproducible and replicable research.
They focused on three main causes for the lack of reproducibility, these included sloppy science, selection and experimental bias, and misconduct. So we had those, and at least I had those in my list, but as we saw there's many other reasons that are less glamorous or less…maybe we say they're more humble reasons for why we might have a problem with reproducibility or replicability. And so I feel like blaming our problems on these three issues is really naive and shortsighted.
And I'd like to think that if you or me were in the room at that colloquium, we perhaps would have come up with different reasons than these three for the problems in reproducibility and replicability. And so a question to think about as we all focus on trying to improve the reproducibility and replicability of our research is, is that research that's perfectly reproducible and perfectly replicable, is it necessarily correct?
Is research that is reproducible and/or replicable necessarily correct?
And I would say no. But I'm optimistic that research that is done well and is described well and is transparent is more likely to be done correctly. If nothing else, it provides an avenue for others to see how sensitive an analysis is to deviations from the pipeline that we propose and to perhaps go back and figure out where we were wrong in our assumptions or incorrect in our data analysis.
Is a result that fails to replicate someone's "fault"?
Following up on that, is a result that fails to replicate someone's "fault"? Can we blame somebody if we can't replicate previous work? I would say, not necessarily. Sure, there's problems of sloppiness and bias and fraud, but we might also be talking about biological phenomenon that we haven't captured.
And it's also important to keep in mind that replication is a product of biological as well as statistical variation, that a small P value does not guarantee the correct result. So as we move forward, a theme that we're going to come back to time and again is the issue of documentation transparency.
And so, I love this meme because it gets to the root of a lot of issues we have with reproducibility and replicability. I'd love to draw this beautiful owl, and so the instructions in the book are how to draw an owl. Draw two circles, draw the rest of the owl. And so, sure those are instructions, those are methods, somebody could perhaps use them to draw an owl.
But it's really not reproducible, it wouldn't tell me much about, say, how to draw a pigeon or a robin. And I believe that there's a reproducibility crisis, if you want to call it that, because our description of methods is pretty lacking. We need to learn how to use existing technology to virtually electronically say, expand our method sections.
Case study
Your lab publishes a paper and gets inundated by emails asking about the nitty gritty of the methods. The trainee that did the study has gone on to a new job.
Discuss this with your PI or other people in your lab...
- Have you ever requested information from an author? What happens?
- Have you ever been pinged for information? What happens?
- What can you do to be proactive to avoid these cases?
- How useful are laboratory notebooks?
- How long are you responsible for maintaining these records?
- How long can you reasonably expect someone to be helpful?
And so I'd like you to think about a case study that is very common and never ceases to amaze me how unprepared I am for this when it happens. That say my lab publishes a paper that's really awesome, it gets a lot of attention, and I start getting emails from people asking about the nitty gritty of how we did things, but not because they want to throw rocks but because they want to do it too.
Unfortunately, because of how peer review works, the training now is long gone, they're off to a new, exciting job doing fun and exciting problems, and they are very slow to answer my emails. I suspect if you talk about this with your PI or other researchers, they'll have this problem as well.
And perhaps you've also been the person to email the PI asking questions. And so what happens when you request this information from the author? And have you ever been on the other side of this, and been asked for information, and what happens? And so I'd like you to just think about these questions and discuss them with your PI, and have a discussion about this, and perhaps motivate a justification for why reproducibility and replicability are important.
And then I'd like you to think also about, what can you do to be proactive to avoid these cases? How do we not have that anxiety when we get these emails from people saying, "What did you do here, I don't understand?" A lot of information is embedded in laboratory notebooks. So how useful are laboratory notebooks when we get these emails, especially as it relates to a computational analysis?
And then, how long are you responsible for maintaining these records? Okay sure, if you get an email of the daily thing is published, you're on the hook. But you know, 3 months later, 6 months later, a year, 5 years, 10 years, 20 years, how long are you responsible for maintaining that? And then, how long can you reasonably expect someone to be helpful?
I would hope that…again the day my paper is published that the trainee is going to be responsive and helpful, but again 5, 10 years later, do I expect them to be helpful? I don't think so, but I don't know when exactly it transitions from expecting someone to be helpful or to have the records, to not having the records or not be helpful.
And so this was a real question, and an interesting thing that we all run into, is getting these emails or getting requests for further information. And so again, discuss this with your PI, discuss this with your lab, what are some of the stories from your lab of when this has happened? And what do you do? What does your PI do? What are their strategies for dealing with this issue?
Philip Bourne baited others into doing this experiment
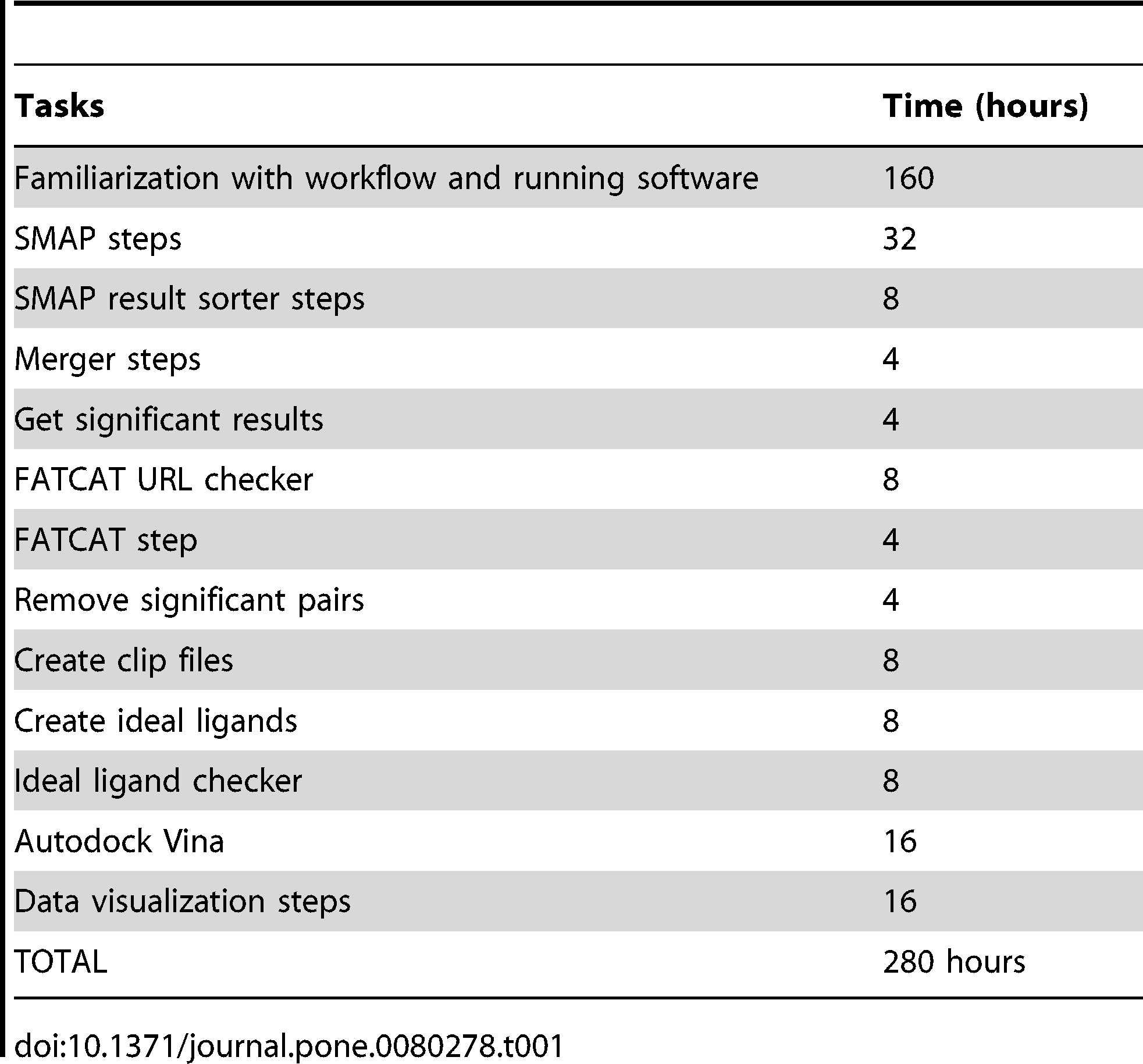
- In 2011, he challenged those attending the Beyond the PDF workshop to reproduce the work performed in his group's 2010 article The Mycobacterium tuberculosis Drugome and Its Polypharmacological Implications
- The result was a detailed analysis of what it would take an author, expert, novice and someone with minimal bioinformatics training to reproduce the analysis
- Training and detailed/automated workflows were seen as critical factors (as well as time and money)
So Philip Bourne baited others into doing this experiment with him. So they published a paper in 2010, looking at the Mycobacterium tuberculosis Drugome, and he encouraged people at a workshop in 2011 to try to reproduce the work in that paper. And Bourne is a thought leader I would consider in the field of bioinformatics, very sensitive to issues of reproducibility.
And they did an audit of how long it would take people at varying stages of their career and familiarity with bioinformatics skills to reproduce a pipeline, to reproduce the results from the paper. And what they found was that, if you had somebody that had pretty basic skills in bioinformatics, that it would take them 160 hours to get up to speed with the workflow and software.
Okay, so that's four 40-hour weeks, just getting up to speed on the workflow. And then it would took another 120 hours to actually implement the workflow. So I'm not convinced that that's…well, it is reproducible. I think it's beyond the level that a lot of us are willing to spend to reproduce somebody's work.
And so it really sheds a lot of light on, what do we mean by reproducible? How much friction if you will do we allow and still say that something is reproducible? If it takes 160 hours, is that reproducible? What knowledge does an individual have to have to be able to reproduce the work? And keep in mind that you're the person doing the work, that knows far more about the work than anybody else ever will.
The Reproducibility Crisis: Bayer
An unspoken rule among early-stage venture capital firms that “at least 50% of published studies, even those in top-tier academic journals, can't be repeated with the same conclusions by an industrial lab"
Prinz et al. 2011. Nature Reviews Drug Discovery
And so, assuming that somebody has the same skill set as you is probably a bad assumption. So, a lot of the issue and discussion around reproducibility crisis comes from two papers that I'd briefly like to mention. The first comes from scientists at Bayer, and they stated that there is an unspoken rule among early-stage venture capital firms, that at least 50% of published studies, even those in top-tier academic journals can't be repeated with the same conclusions by an industrial lab.
Okay, so say Bayer sees a really cool results from an academic researcher, they want to perhaps convert that into a product for selling commercially, but they find that at least 50% of those preclinical studies just cannot be repeated. And so, in this review they talk about…I think there's about 67 or so, maybe 70 studies that they had tried to reproduce or replicate at Bayer.
The Reproducibility Crisis: Bayer
An unspoken rule among early-stage venture capital firms that “at least 50% of published studies, even those in top-tier academic journals, can't be repeated with the same conclusions by an industrial lab"
Prinz et al. 2011. Nature Reviews Drug Discovery
And so, assuming that somebody has the same skill set as you is probably a bad assumption. So, a lot of the issue and discussion around reproducibility crisis comes from two papers that I'd briefly like to mention. The first comes from scientists at Bayer, and they stated that there is an unspoken rule among early-stage venture capital firms, that at least 50% of published studies, even those in top-tier academic journals can't be repeated with the same conclusions by an industrial lab.
Okay, so say Bayer sees a really cool results from an academic researcher, they want to perhaps convert that into a product for selling commercially, but they find that at least 50% of those preclinical studies just cannot be repeated. And so, in this review they talk about…I think there's about 67 or so, maybe 70 studies that they had tried to reproduce or replicate at Bayer.
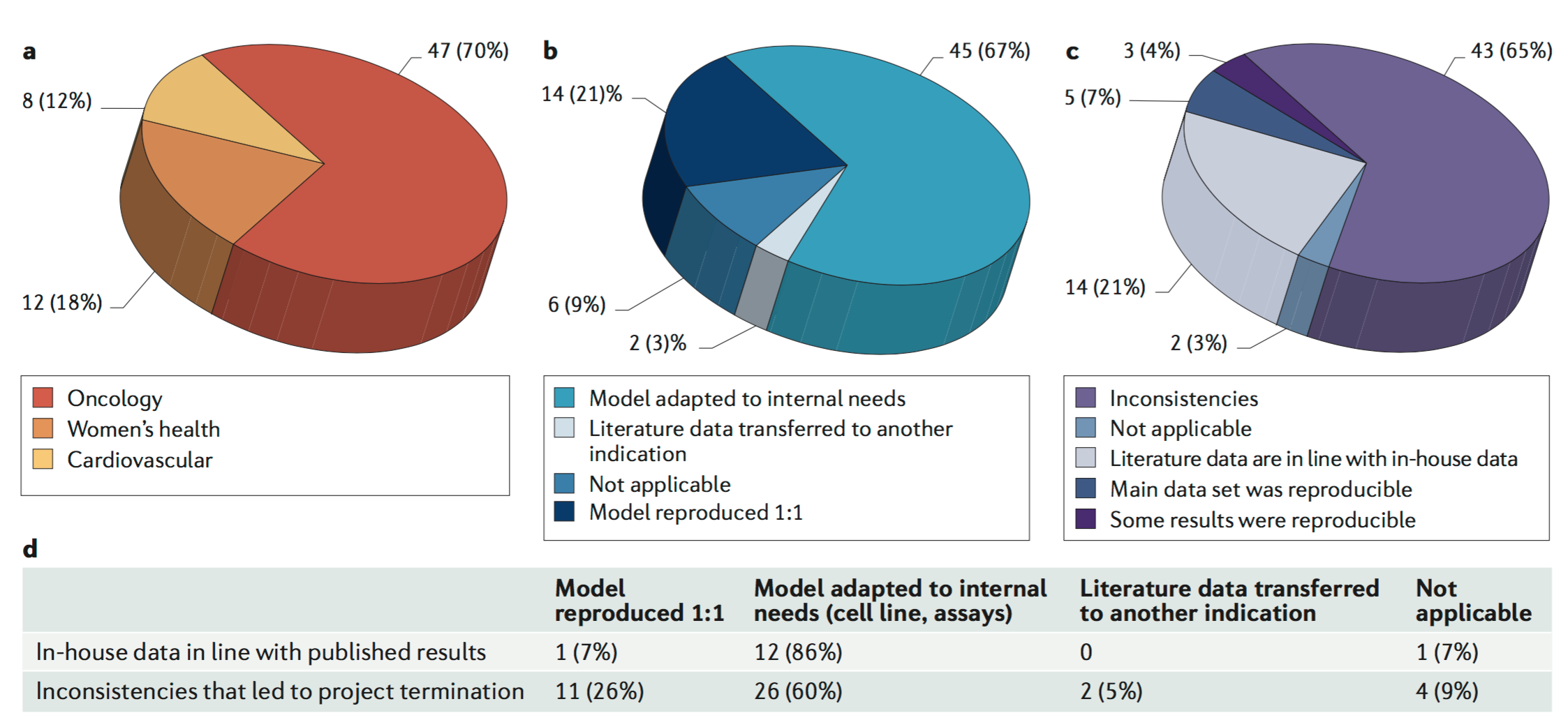
And what they found was that, there were inconsistencies in about 65% of the studies they studied, and that only about 4% were some of the results reproducible. And in about 7% of the studies where the main dataset was reproducible. And so this is a…this seems like a problem, where a large fraction of the studies that they were testing just could not be reproduced.
Now, there are subtleties to this because they were not doing exact reproductions, these would perhaps by our rubric considered to be replications, or perhaps they were using different mouse models or they were using a different setup to fit what they were doing in their labs, but still it points to an issue.
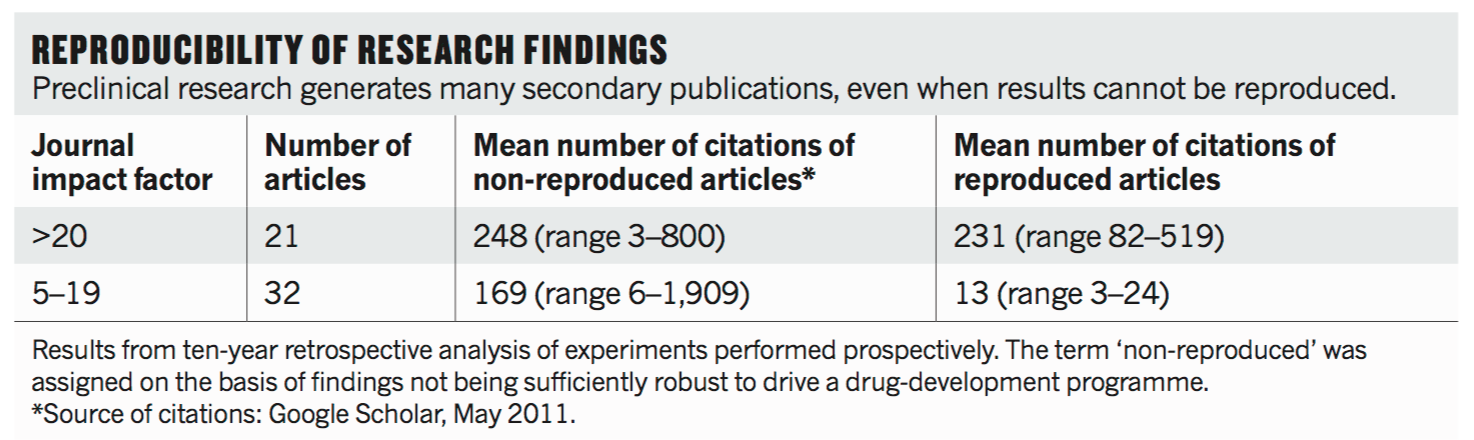
The next study was published by Amgen by Begley & Ellis, and they found that regardless of the impact factor of the journal that the original result was published in, that they found many of those studies also were not reproducible or replicable.
And furthermore they pointed out that a number of other studies would come on after the initial result, trying to follow up on specific points or building off of the earlier work to take the work further. And so their point is that there's a lot of wasted time and resources in trying to build upon flawed science.
The Reproducibility Crisis: Amgen
Cancer researchers must be more rigorous in their approach to preclinical studies. Given the inherent difficulties of mimicking the human micro-environment in preclinical research, reviewers and editors should demand greater thoroughness
The next study was published by Amgen by Begley & Ellis, and they found that regardless of the impact factor of the journal that the original result was published in, that they found many of those studies also were not reproducible or replicable.
And furthermore they pointed out that a number of other studies would come on after the initial result, trying to follow up on specific points or building off of the earlier work to take the work further. And so their point is that there's a lot of wasted time and resources in trying to build upon flawed science.
And so they state that cancer researchers must be more rigorous in their approach to preclinical studies. Given the inherent difficulties of mimicking the human micro-environment in preclinical research, researchers and editors should demand greater thoroughness. So that was a very negative, I would say "gotcha science" type of attitude.
Inconvenient Truth: Neither study did anything to demonstrate that their work could be reproduced
And so, but there's something that maybe you've noticed if you've looked at these articles, is that neither study did anything to demonstrate that their work as demonstrated in these studies, these reviews could be reproduced, that the very thin details as to what they did, how they tried to reproduce or replicate these experiments.
And so you maybe want to pause and think deeply about how much we want to focus on these two reviews. But at the same time there is a general sense that science is hard, science has gotten very complicated in the last decades, and that we are not doing a good job of ensuring the reproducibility and replicability of our own research.
NIH's Response: Technical Factors
- Publications rarely report basic elements of experimental design
- Blinding, Randomization, Replication
- Sample-size calculation
- Effect of sex differences
- Underlying data rarely made publicly available
- Restrictions on lengths of methods sections
- Assume reader is at an idealized level of expertise
And so to that, Collins & Tabak published an editorial in Nature in 2014 talking about NIH's response to the so called reproducibility crisis. And they point out a handful of problems that they then wanted to follow up on in policy changes at NIH. So they talk about publications that rarely report basic elements of experimental design, things like blinding, randomization, replication, sample size, effect of sex differences, now the underlying data really made available.
NIH's Response: Social Factors
- Failure to publish negative effects (i.e. file drawer problem)
- "Some scientists reputedly use a 'secret sauce' to make their experiments work"
- Perverse incentives to publish and hype striking results
- Impact Factor Mania
There's a problem because there's restrictions on the length of method sections. And then, as we saw with the Bourne paper, they assume that the reader has an idealized level of expertise, that's just not practical. So NIH has focused on a lot of these social factors. And so the idea that we fail to publish negative results, this is commonly called the file drawer problem, that we stick our negative results into a file drawer and never report them again.
Some scientists they say reputedly use a "secret sauce" to make their experiments work, this was a problem, lack of transparency. There's also perverse incentives to publish and hype striking results, and this ties in with the idea of the impact factor mania where people are very focused on getting their work published into a journal with a high impact factor.
That the ability to secure a K Award or to get tenure or promotion is tied to the number of papers that you have in journals with very high impact factors. And as I said, NIH has gone out of the way now to go ahead and think about new guidelines that they have imposed to improve the reproducibility and replicability of our research, and thinking about, how do we justify the premise?
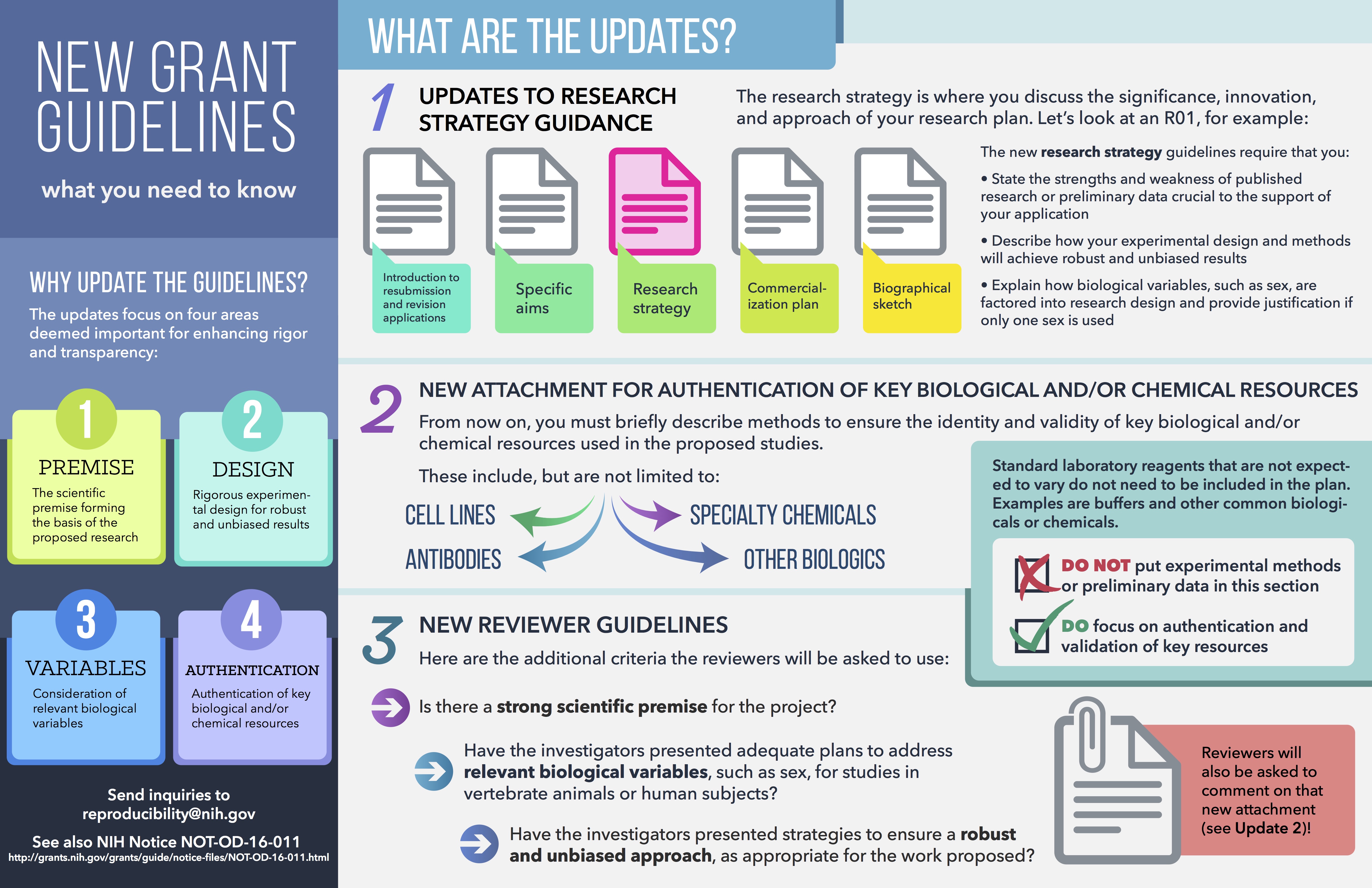
Experimental design issues. What are biologically relevant variables? How do we authenticate our reagents to know that they are what we think they are? There's been a big push by NIH to improve the reproducibility and replicability of ongoing research, and much of these efforts have focused on what we are calling replicability.
This tutorial series was funded in part by an R25 that I received from NIH dealing with reproducibility in the area of microbiome research.
Thinking about the microbiome world, Jacques Ravel and Eric Wommack published an editorial in Microbiome called "All hail reproducibility in microbiome research."
This was one of the assigned readings before this tutorial. So I want to ask what your initial thoughts were about this, what do you think? One of the comments that jumped out at me was when they said, it is no mistake that the best documented code turns out to be more frequently used by microbiome researchers. That if you go out of your way to make your data available, you make your code available, you document it, you make it clear what you've done, it is going to get cited, it is going to be widely used.
I have certainly experienced that with our Mothur software package, that by making it as open and transparent and well documented as we can, we have gotten a large number of citations because we have engaged in those practices. So what I'd like you to do is take the next four or five minutes and read back through this editorial, and see if you can identify three or four different technologies or platforms that the authors point to for improving reproducibility of microbiome research.
So go ahead and hit pause, and once you've done that go ahead and come back. All right, so they focus on several tools related to data accessibility, including depositing sequence data in the Sequence Read Archive, as well as in dbGap.
Other data we might put into a website called Figshare as a repository for data. Using a metadata standard called min marks. Writing our code and our workflow together using tools like the Project Jupiter, Notebooks which they call in this editorial IPython, there's been a switch in name since this was published, as well as knitter documents in the R environment.
And then finally they also talked about using version control tools, including software like Git, as well as a website built on top of Git called GitHub. And so as we go through these series of tutorials, we're going to describe how we would use these different tools to improve the reproducibility of our own research.
We're also going to go considerably further than Ravel and Wommack went in this editorial to further improve the reproducibility of our research.
Threats to reproducibility & replicability of microbiome research
So if we think about microbiome research in particular, we might think about different threats to reproducibility and replicability. And so these are things that I want to build on beyond the lists that we came up with earlier.
Threats to reproducibility & replicability of microbiome research
- Lack of standard methods
- Accessibility of data
- Different populations
- Complex and lengthy data analysis
- Variation in mouse colonies
- Contaminants in low biomass samples
- Sampling artifacts
So if we think about microbiome research in particular, we might think about different threats to reproducibility and replicability. And so these are things that I want to build on beyond the lists that we came up with earlier.
And so one of the issues is a lack of standard methods. And this is a very thorny issue with a lot of people, myself included, because we pick our methods to answer specific questions that are relevant to us today. Like I use the variable region I use to sequence the 16S rRNA gene because I've got a specific set of questions.
Well, if you're studying a different set of questions or in a different part of the body, you might want to use a different region. That's going to make it really difficult to compare results in a meaningful way. The accessibility of data is still a big problem, not everybody is depositing data in the SRA, not everybody is providing metadata to actually make that data useful.
We all use different populations, that's kind of the beauty of the microbiome field right now, is that there are people asking the same question in different cohorts. We also have really complicated and lengthy data analysis pipelines. You know there's many steps, there's many parameters, many variables we might use, different options for different databases we might use.
It's complicated, it's lengthy. One of the things that drives me nuts is reading a manuscript that says, "We analyzed our 16S rRNA gene data using Mothur." And that's it, they don't say anything about how they used Mothur. I know as the Mothur developer that there's thousands of ways you could use Muther to analyze your data, and so we need greater clarity to help us through this complexity and lengthiness of our data analysis pipelines.
There's variation in our mouse colonies. If you look at the gut microbiota of mice from Jackson versus Taconic, versus those in the breeding facility here at the University of Michigan, we're going to see big differences. Those differences can cause differences in the phenotypes we're interested in. We also know that there are contaminants that show up from our reagents when we're sequencing low biomass samples, this has raised a lot of questions in regards to whether or not there's a microbiome associated with the placenta or with lungs.
We also know that there are sampling artifacts, where how we store our samples, what we do with our samples, the size of our samples can all introduce artifacts that are going to impact our downstream results.
Case study
Consider the GitHub repository that accompanied the paper by Meadow et al. described by Ravel & Wommack
- How accessible is the code?
- What do you need to know to make sense of the repository?
- Is it well documented?
- How well organized is the repository?
- How long did it take you to find the code for the figures?
What I'd like you to do next is go back to that Ravel and Wommack editorial, and look at the paper by Meadows et al, as well as the GitHub repository that they cite in their paper.
And so I'm going to pull that up for you here. And so this is the GitHub site that has the repository for Meadows et al. And what I'd like you to do is take some time to see how accessible is the code.
Okay, the code is here, how accessible is it? Can you make heads or tails of what's going on? What do you need to know to make sense of the repository? Is it structured? Is it organized in a way that you can understand what's going on? Is it well documented? How long did it take you to find the code for the figures in the paper?
So go ahead and go through this, and think about the reproducibility or how this helps improve the reproducibility of the paper from Meadows et al. Great. So I really…my hat…I tip my hat to Meadows et al because they put this up at a time when very few people were thinking about how to make their work more reproducible.
And so I really credit them with doing a good job of making their code accessible. That being said, I think there's still a number of points that they could improve upon to make the work more reproducible, more transparent, more accessible. So first of all, the sequenced data that they published was deposited in the Figshare and not into the Sequence Read Archive.
There have been times in the past where the Sequence Read Archive has been really difficult to work with for submitting 16S rRNA gene data, and I suspect that's why they ultimately put it into Figshare. It's not a big project, but still the code isn't very well organized. It doesn't jump out at me where the data are, where the code is, where the output is.
The good thing is that the RMD file provides the narrative to explain what's happening and why they made different decisions. And the code for the figure is in the littlesurfaces.rmd file, and it's rendered in the littesurfaces.md file.
And so it's there, I could certainly find areas to improve upon. But again as a first step, and one of the first studies that really dug into making their work more reproducible, I think they did a great job. If we come forward in time a bit, we might think about more recent microbiome papers and how well they've gone out of their way to make their code, their analysis more reproducible.
Critique a recent microbiome paper
Basic checklist
- How difficult it be to regenerate a P-value or a figure?
- Where are the raw data?
- Is the code available?
- How long is the Methods section?
Some possible examples from the Schloss Lab...
And so I'm taking three repositories for my own research group for three papers over the last few years. I don't consider these to be perfect, I think if you go through from Zackular, to Baxter, to Sze, you'll see differences and changes in how my group has approached, ensuring the reproducibility of our data analysis pipelines.
And so something that we can use as a checklist to assess a recent microbiome paper might be how difficult is it to regenerate a P value or a figure? Could we go all the way back to raw data to regenerate that P value or a figure? Where are the raw data? Is the code available? How long is the method section?
Do they do that thing where they say, "We used Mothur?" Or do they tell you about the databases they used? Do they cite the R packages that they used? So I'd really encourage you to go through these three examples that I think are pretty good and biased of course, but at the same time I know we've evolved in how we approach these types of repositories, and our approach to ensuring that things are reproducible.
So go ahead and take a few minutes to check out those repositories. Great. So I hope you enjoyed looking at those. And the repository that we see for Sze & Schloss is more along the lines of what we're going to have as an output from our work at the end of this series of tutorials.
Why is reproducibility/replicability important?
So why is reproducibility and replicability important?
Why is reproducibility/replicability important?
- To make sure that the results are correct and/or generalizable
So why is reproducibility and replicability important?
Perhaps we think that's obvious, well obviously we want to make sure that our results are correct and are generalizable. And I think this is where most people stop, when they're thinking about reproducibility and replicability, they're thinking about correctness.
Why is reproducibility/replicability important?
- To make sure that the results are correct and/or generalizable
- So that others can build off of earlier work
So why is reproducibility and replicability important?
Perhaps we think that's obvious, well obviously we want to make sure that our results are correct and are generalizable. And I think this is where most people stop, when they're thinking about reproducibility and replicability, they're thinking about correctness.
And as we said earlier, just because something is reproducible doesn't necessarily mean it's correct. I'm also interested in making sure that others can build off of my work. And I also want them to take my work and repurpose the materials and methods to do something different.
Why is reproducibility/replicability important?
- To make sure that the results are correct and/or generalizable
- So that others can build off of earlier work
- To enable others to repurpose materials and methods
So why is reproducibility and replicability important?
Perhaps we think that's obvious, well obviously we want to make sure that our results are correct and are generalizable. And I think this is where most people stop, when they're thinking about reproducibility and replicability, they're thinking about correctness.
And as we said earlier, just because something is reproducible doesn't necessarily mean it's correct. I'm also interested in making sure that others can build off of my work. And I also want them to take my work and repurpose the materials and methods to do something different.
So we've done a lot in the area of finding signatures of colorectal cancer using 16S rRNA gene sequencing. Well, if somebody publishes a new study doing the same type of thing and they don't use my data, that's a bummer. I mean, we invested a lot of time and effort and money into generating and analyzing those data, It'd be a shame that others wouldn't use those data to build upon for their own work.
Similarly, if people have looked at our code and found it useful to answer a question for a certain set of subjects or experiments, I would hope they'd find it useful for their own set of conditions and experiments and populations. So ultimately, I think of reproducible research as being a form of preventative medicine, and this is a phrase that was coined by Jeff Leak and Roger Peng in that editorial I mentioned at the beginning of this tutorial.
Preventative medicine
- Too much emphasis on "gotcha science"
- Research done in a manner to maximize reproducibility...
- makes life easier for you in the long run
- instills more confidence by others
- is easier for others to build from
There's way too much emphasis on "gotcha science,"way too much emphasis on the Amgen and Bayer papers. Research that's done in a manner to maximize reproducibility is going to make life easier for you in the long run. So you get that email from someone's that's excited about your work, if you could send them to the repository that has all your code and it's well documented, wouldn't that be amazing?
It's going to instill more confidence in others, and then it's going to be easier for others to build from. And so really think about this as a form of preventative medicine, as a way of fostering collaboration with others going forward. And so as we think about collaboration, your collaborators need to be able to reproduce your research.
Who must be able to reproduce your research?

And so who has to be able to reproduce your research? I told you this at the end of the last tutorial, but you, you have to be able to reproduce your own research. And think about that in six months from now, and think that currently you no longer has access to email, how would you write for yourself six months from now knowing that you couldn't get in a time machine or email old you to figure out what you were doing? So take care of yourself, be preemptive in how you're thinking about reproducibility.
Who must be able to reproduce your research?


And so who has to be able to reproduce your research? I told you this at the end of the last tutorial, but you, you have to be able to reproduce your own research. And think about that in six months from now, and think that currently you no longer has access to email, how would you write for yourself six months from now knowing that you couldn't get in a time machine or email old you to figure out what you were doing? So take care of yourself, be preemptive in how you're thinking about reproducibility.
Second, your PI is an important collaborator, they need to be able to reproduce what you're doing, because at some point you're hopefully going to graduate and move on to greener pastures, and your PI is going to be left behind to figure out what the heck you did and how they can communicate that to other researchers who want to build upon your work but also their own work.
Where we're going
- Documentation
- Keep raw data raw
- Data organization as a form of documentation
- Script everything
- Don't repeat yourself (DRY)
- Automation
- Collaboration
- Transparency
So where are we going with this series of tutorials? So we're going to spend a lot of time in the next session talking about documentation in terms of text-based documentation, but also in future talks about data organization as a form of documentation, using code as a form of documentation, and thinking about automation as a form of documentation.
That if we can tell the computer how to run the analysis in an automated way, well then we need to convert from computer speak, from code to human texts to understand what's going on. And along the way as we learn some best practices like keeping our raw data raw as much as possible and not touching it with our hands or with our cursor, not repeating our self with our code, and then using tools that enable collaboration and transparency.
Exercises
- How difficult would it be for you to regenerate any of the figures or p-values from a paper that your lab published 5 yrs ago?
- What would be the most important thing to improve for your group's next paper?
- What percentage of papers published in your field do you think are reproducible? Replicable? Why?
- Create an ordered list of the broad groups of people that need to be able to reproduce your work
So in closing, I'd like you to spend some time thinking about a series of questions. Then again, think about discussing these with your PI, with your research group, with your friends. So first, how difficult would it be for you to regenerate any of the figures or P values from a paper that your lab published five years ago?
So if you open up a paper from five years ago and go to figure two, how difficult would it be for you or one of your current lab mates to figure out how to regenerate that plot, that figure? For your next paper, what's the most important thing that you can do to improve the reproducibility of that data analysis?
And so Amgen and Bayer said they thought it was about 50%, but what percent of papers published in your field do you think are reproducible, replicable? And then why do you come up with those numbers? And then finally, building out upon your two important collaborators of yourself and your PI, what other broad groups of people need to be able to reproduce or replicate your work?
And think about them in terms of their own skill sets and their own knowledge. And this gets back to the idea of the Bourne paper where it took somebody with decent bioinformatic skills a long period of time to get up to speed with the analysis that was done in a previous paper.
Wonderful. Hopefully you found today's discussion helpful in getting you to think about your own reproducible research practices, and where there might be gaps in your practices. At the same time, I hope you can have a greater appreciation that everybody has gaps in their practices. The stuff is really hard. Please be sure to take the time to engage those in your research group, your lab mates, your PIs, and those around your lab to discuss the material and questions that came up in today's tutorial.
Next time, we'll start to develop more practical steps to greater reproducibility, we'll slowly be ratcheting up the technical material in the next tutorial. Until then, have fun engaging these questions and having discussions with those around you and those you're doing research with. I think you'll find it to be really useful, really illuminating, and really helping to point the direction as you grow in your own ability to do reproducible and replicable research.

