Session 6
Topics
- Finding inspiration
- Specifying variable types with
read_csv selecting-ing columns from a data frame- Renaming columns in a data frame
Finding inspiration
I’ve frequently heard old timers who live in Southeastern Michigan tell me that we used to get a lot more snow, ponds used to stay frozen longer, and it’s never been as warm as it has been in the last few summers. There’s a lot of anecdotal evidence from these farmers that the climate is changing.
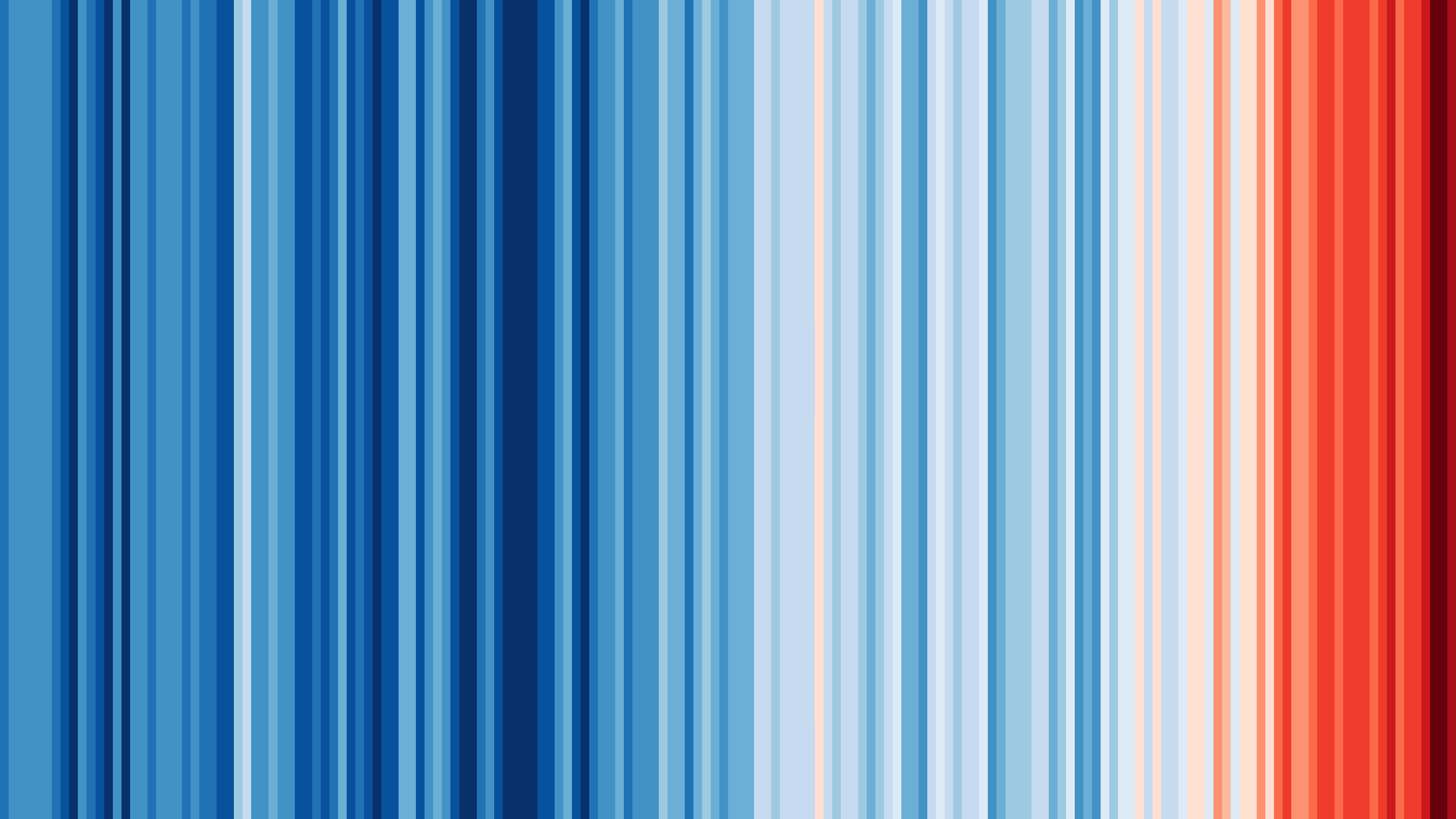
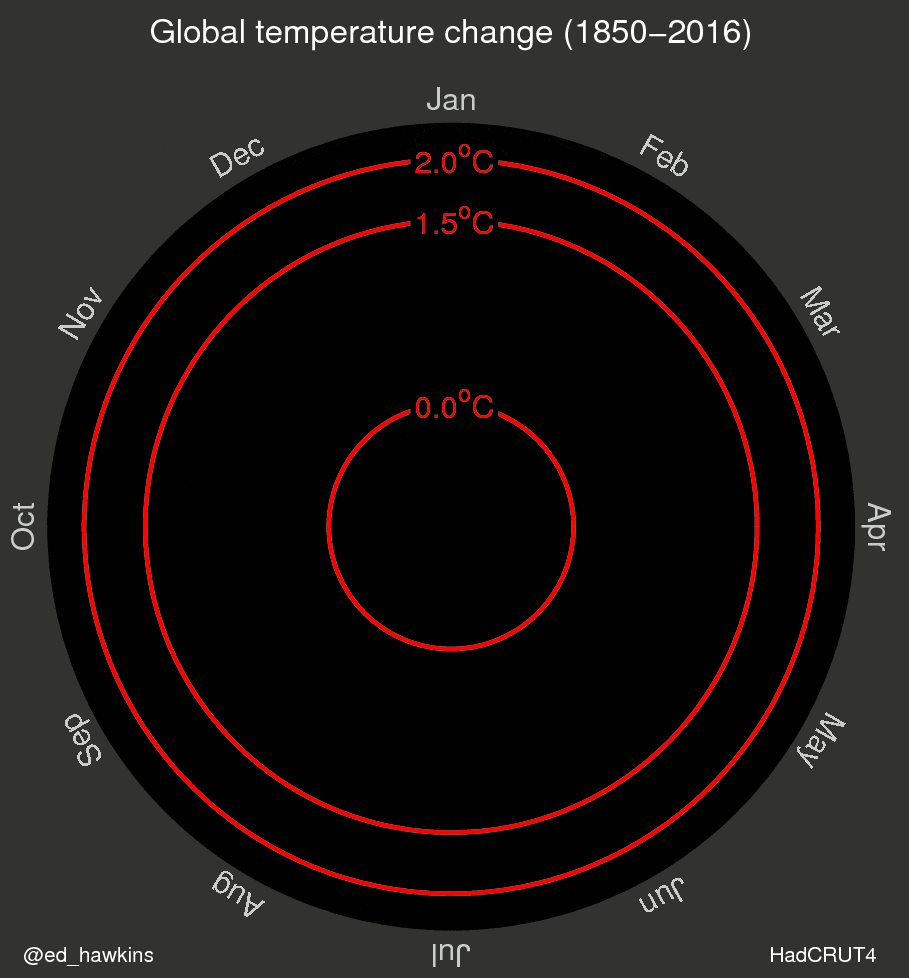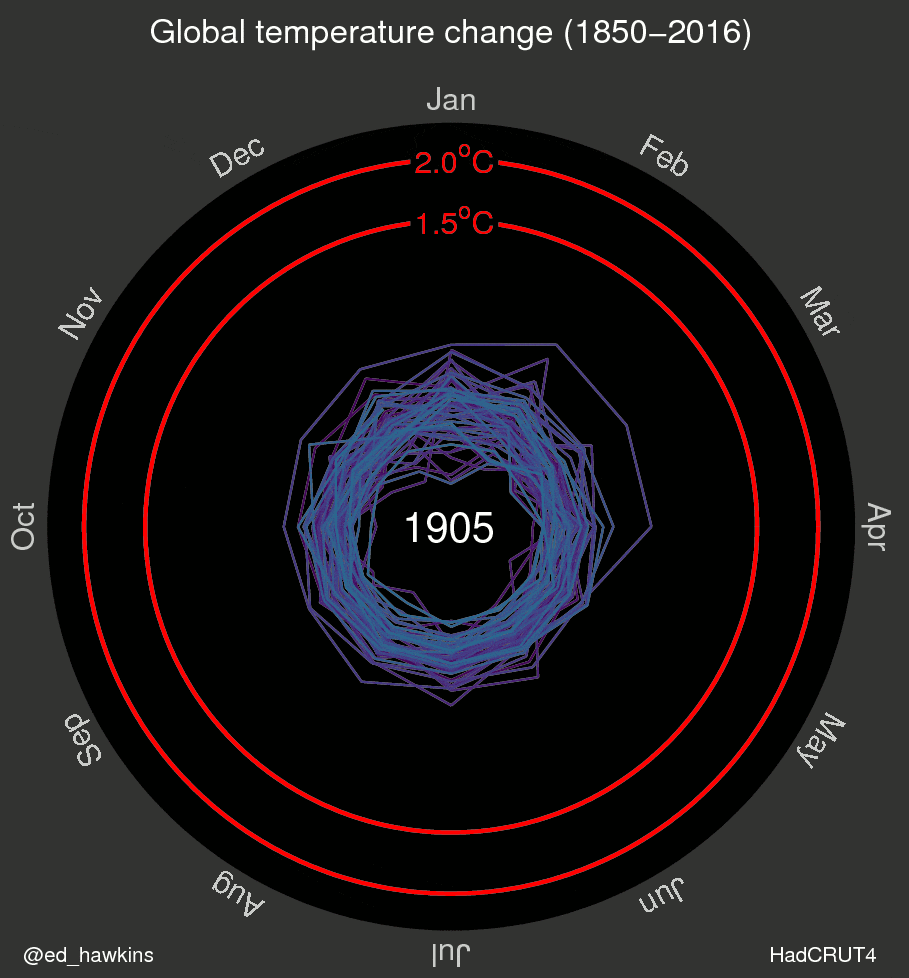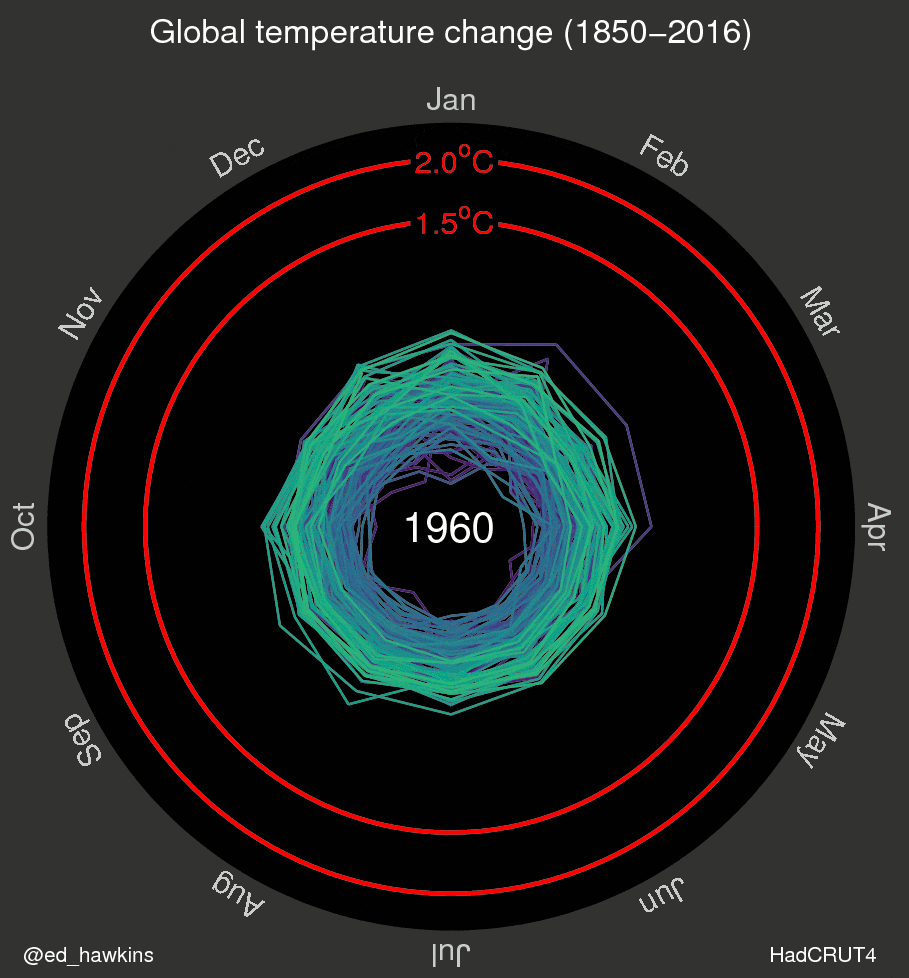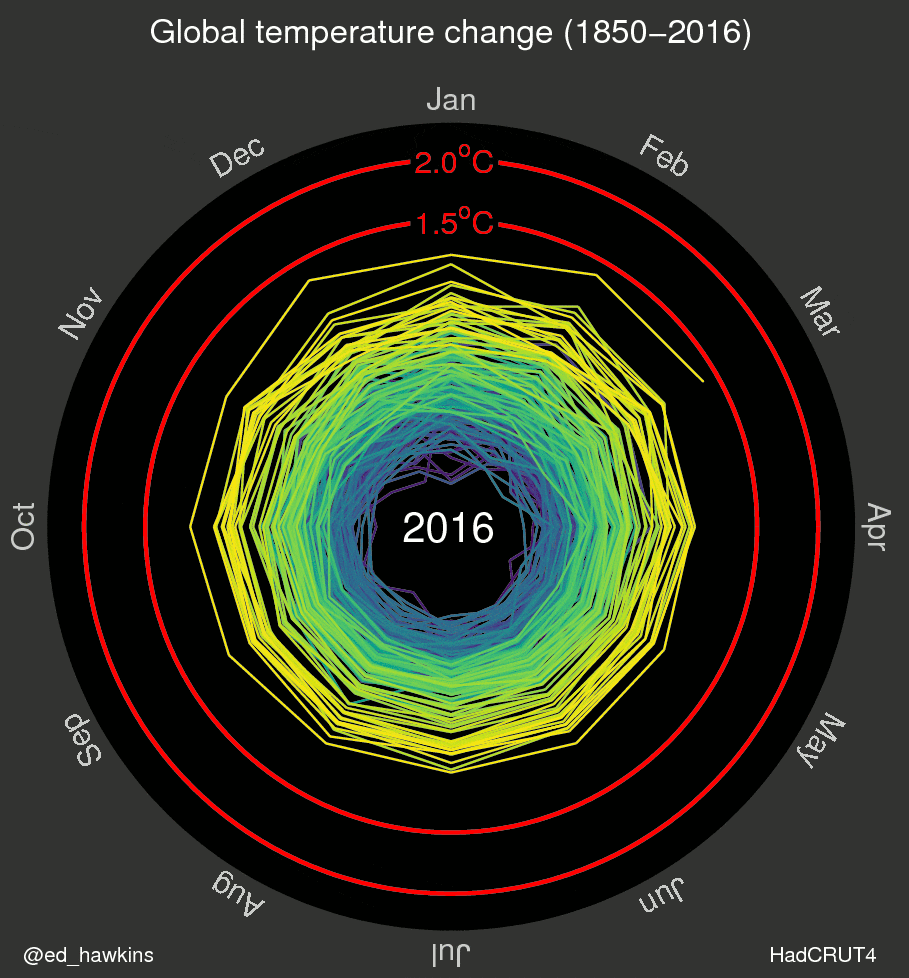
I’ve also been inspired by a few recent data visualizations and wondered what they would look like for various places I’ve lived. These popular visuals were generated using global average temperatures dating back to about 1850
It’s important to keep in mind that many visualizations are for a global average and that some places may be cooling while others are warming. I’ve been curious if we can see that in the data. For the next few sessions we’re going to switch to a different dataset that includes weather observations from Ann Arbor, MI from October 1891 through the beginning of April 2020. Let’s take a look at what has been happening in Ann Arbor over the past 130 years.
If you know the identifier for your local weather recording station, you can obtain data near where you live from NOAA who maintains the Daily Global Historical Climatology Network. My local weather station with the most data is located on the north side of the University of Michigan campus and has the code “USC00200230”. The data we’ll use is from that weather station and includes temperature and precipitation data going back to 1891. You can get an overview of the data available at the station’s summary page. I’ve put a comma-separated values (CSV) version of the file in your noaa directory.
Specifying variable types with read_csv
We’ll start by loading the tidyverse and reading in the data frame for my local weather station.
library(tidyverse)
read_csv('noaa/USC00200230.csv')
##
## ── Column specification ────────────────────────────────────────────────────────────────────────────
## cols(
## STATION = col_character(),
## NAME = col_character(),
## DATE = col_date(format = ""),
## DAPR = col_logical(),
## DASF = col_logical(),
## MDPR = col_logical(),
## MDSF = col_logical(),
## PRCP = col_double(),
## SNOW = col_double(),
## SNWD = col_logical(),
## TMAX = col_double(),
## TMIN = col_double(),
## TOBS = col_logical()
## )
## Warning: 70075 parsing failures.
## row col expected actual file
## 1889 SNWD 1/0/T/F/TRUE/FALSE 0.0 'noaa/USC00200230.csv'
## 1890 SNWD 1/0/T/F/TRUE/FALSE 0.0 'noaa/USC00200230.csv'
## 1891 SNWD 1/0/T/F/TRUE/FALSE 0.0 'noaa/USC00200230.csv'
## 1892 SNWD 1/0/T/F/TRUE/FALSE 0.0 'noaa/USC00200230.csv'
## 1893 SNWD 1/0/T/F/TRUE/FALSE 0.0 'noaa/USC00200230.csv'
## .... .... .................. ...... ......................
## See problems(...) for more details.
## # A tibble: 46,650 x 13
## STATION NAME DATE DAPR DASF MDPR MDSF PRCP SNOW SNWD TMAX
## <chr> <chr> <date> <lgl> <lgl> <lgl> <lgl> <dbl> <dbl> <lgl> <dbl>
## 1 USC002… ANN … 1891-10-01 NA NA NA NA NA NA NA 20.6
## 2 USC002… ANN … 1891-10-02 NA NA NA NA NA NA NA 26.7
## 3 USC002… ANN … 1891-10-03 NA NA NA NA NA NA NA 26.1
## 4 USC002… ANN … 1891-10-04 NA NA NA NA 7.6 NA NA 22.8
## 5 USC002… ANN … 1891-10-05 NA NA NA NA NA NA NA 13.9
## 6 USC002… ANN … 1891-10-06 NA NA NA NA NA NA NA 14.4
## 7 USC002… ANN … 1891-10-07 NA NA NA NA 4.1 NA NA 10.6
## 8 USC002… ANN … 1891-10-08 NA NA NA NA NA NA NA 13.9
## 9 USC002… ANN … 1891-10-09 NA NA NA NA NA NA NA 15
## 10 USC002… ANN … 1891-10-10 NA NA NA NA NA NA NA 16.7
## # … with 46,640 more rows, and 2 more variables: TMIN <dbl>, TOBS <lgl>
Running read_csv brings up a warning message that there were “70075 parsing errors”. If you look right above that error message, you’ll see that read_csv tells us that Parsed with column specification… This means that read_csv guessed the type of data that was in each column. For example, it’s assuming that SNWD is a logical. If you look at the output from ?read_csv you’ll learn that the function uses the first 1000 columns of the data to guess the type of data. Looking at the Daily Summaries Station Details webpage for this station, click on the “Precipitation” link under the “Available Data Types” header. To the right, you’ll see that SNWD refers to “Snow Depth”. The station didn’t start collecting those data until 1896. The warning we’re seeing is because it has only seen NA (i.e. missing data, which is a logical) in the first 1000 rows. That read_csv can guess is normally a nice feature, but in this case it’s a bit of a nuisance. We can either turn it off, tell read_csv to use more data to guess the column type, or tell read_csv the actual column types. Let’s see how we’d do each
read_csv('noaa/USC00200230.csv', guess_max = 10000)
##
## ── Column specification ────────────────────────────────────────────────────────────────────────────
## cols(
## STATION = col_character(),
## NAME = col_character(),
## DATE = col_date(format = ""),
## DAPR = col_logical(),
## DASF = col_double(),
## MDPR = col_logical(),
## MDSF = col_double(),
## PRCP = col_double(),
## SNOW = col_double(),
## SNWD = col_double(),
## TMAX = col_double(),
## TMIN = col_double(),
## TOBS = col_logical()
## )
## Warning: 33801 parsing failures.
## row col expected actual file
## 12510 TOBS 1/0/T/F/TRUE/FALSE 1.1 'noaa/USC00200230.csv'
## 12511 TOBS 1/0/T/F/TRUE/FALSE 1.1 'noaa/USC00200230.csv'
## 12512 TOBS 1/0/T/F/TRUE/FALSE 0.6 'noaa/USC00200230.csv'
## 12513 TOBS 1/0/T/F/TRUE/FALSE 3.9 'noaa/USC00200230.csv'
## 12514 TOBS 1/0/T/F/TRUE/FALSE 2.8 'noaa/USC00200230.csv'
## ..... .... .................. ...... ......................
## See problems(...) for more details.
## # A tibble: 46,650 x 13
## STATION NAME DATE DAPR DASF MDPR MDSF PRCP SNOW SNWD TMAX
## <chr> <chr> <date> <lgl> <dbl> <lgl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 USC002… ANN … 1891-10-01 NA NA NA NA NA NA NA 20.6
## 2 USC002… ANN … 1891-10-02 NA NA NA NA NA NA NA 26.7
## 3 USC002… ANN … 1891-10-03 NA NA NA NA NA NA NA 26.1
## 4 USC002… ANN … 1891-10-04 NA NA NA NA 7.6 NA NA 22.8
## 5 USC002… ANN … 1891-10-05 NA NA NA NA NA NA NA 13.9
## 6 USC002… ANN … 1891-10-06 NA NA NA NA NA NA NA 14.4
## 7 USC002… ANN … 1891-10-07 NA NA NA NA 4.1 NA NA 10.6
## 8 USC002… ANN … 1891-10-08 NA NA NA NA NA NA NA 13.9
## 9 USC002… ANN … 1891-10-09 NA NA NA NA NA NA NA 15
## 10 USC002… ANN … 1891-10-10 NA NA NA NA NA NA NA 16.7
## # … with 46,640 more rows, and 2 more variables: TMIN <dbl>, TOBS <lgl>
We see that SNWD is now a double or numeric variable. We can also turn off the guessing by telling read_csv to use all of the rows to figure out how to parse the columns….
read_csv('noaa/USC00200230.csv', guess_max = Inf)
## Warning: `guess_max` is a very large value, setting to `21474836` to avoid
## exhausting memory
##
## ── Column specification ────────────────────────────────────────────────────────────────────────────
## cols(
## STATION = col_character(),
## NAME = col_character(),
## DATE = col_date(format = ""),
## DAPR = col_double(),
## DASF = col_double(),
## MDPR = col_double(),
## MDSF = col_double(),
## PRCP = col_double(),
## SNOW = col_double(),
## SNWD = col_double(),
## TMAX = col_double(),
## TMIN = col_double(),
## TOBS = col_double()
## )
## # A tibble: 46,650 x 13
## STATION NAME DATE DAPR DASF MDPR MDSF PRCP SNOW SNWD TMAX
## <chr> <chr> <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 USC002… ANN … 1891-10-01 NA NA NA NA NA NA NA 20.6
## 2 USC002… ANN … 1891-10-02 NA NA NA NA NA NA NA 26.7
## 3 USC002… ANN … 1891-10-03 NA NA NA NA NA NA NA 26.1
## 4 USC002… ANN … 1891-10-04 NA NA NA NA 7.6 NA NA 22.8
## 5 USC002… ANN … 1891-10-05 NA NA NA NA NA NA NA 13.9
## 6 USC002… ANN … 1891-10-06 NA NA NA NA NA NA NA 14.4
## 7 USC002… ANN … 1891-10-07 NA NA NA NA 4.1 NA NA 10.6
## 8 USC002… ANN … 1891-10-08 NA NA NA NA NA NA NA 13.9
## 9 USC002… ANN … 1891-10-09 NA NA NA NA NA NA NA 15
## 10 USC002… ANN … 1891-10-10 NA NA NA NA NA NA NA 16.7
## # … with 46,640 more rows, and 2 more variables: TMIN <dbl>, TOBS <dbl>
The only difference we see between these two outputs is pretty subtle - the type for TOBS changed from a logical to a double. If you go back to the station’s summary webpage and click on the “Air Temperature” link, you’ll see that TOBS (Temperature at the time of observation) was first collected in 1926, about 35 years or 12775 days into the dataset. That’s beyond the 10,000 rows or days we used with guess_max = 10000.
Perhaps the best way of setting the variable types is to set them manually using the col_types argument. We can do this a few different ways, but the easiest is to create a string where each letter in the string indicates the type of data in each column. Character data can be represented with a “c”, integer data (counts) with an “i”, double (i.e. decimal) data with a “d”, any numeric data with an “n”, logical data with a “l”, date data with a “D”, time data with a “t”, date-time data with a “T”, and you can tell read_csv to guess the column with a “?”. You can also tell read_csv to skip the column with a “_”. If you forget this (and I do!) you can find the codes in the col_types section of the ?read_csv documentation. Applying this knowledge along with the information on the summary webpage we can do the following…
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd")
## # A tibble: 46,650 x 13
## STATION NAME DATE DAPR DASF MDPR MDSF PRCP SNOW SNWD TMAX
## <chr> <chr> <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 USC002… ANN … 1891-10-01 NA NA NA NA NA NA NA 20.6
## 2 USC002… ANN … 1891-10-02 NA NA NA NA NA NA NA 26.7
## 3 USC002… ANN … 1891-10-03 NA NA NA NA NA NA NA 26.1
## 4 USC002… ANN … 1891-10-04 NA NA NA NA 7.6 NA NA 22.8
## 5 USC002… ANN … 1891-10-05 NA NA NA NA NA NA NA 13.9
## 6 USC002… ANN … 1891-10-06 NA NA NA NA NA NA NA 14.4
## 7 USC002… ANN … 1891-10-07 NA NA NA NA 4.1 NA NA 10.6
## 8 USC002… ANN … 1891-10-08 NA NA NA NA NA NA NA 13.9
## 9 USC002… ANN … 1891-10-09 NA NA NA NA NA NA NA 15
## 10 USC002… ANN … 1891-10-10 NA NA NA NA NA NA NA 16.7
## # … with 46,640 more rows, and 2 more variables: TMIN <dbl>, TOBS <dbl>
We saw a different syntax for col_types when we were working with the Project Tycho data. You’ll recall that we had
read_csv("project_tycho/US.23502006.csv",
col_type=cols(PartOfCumulativeCountSeries = col_logical()))
Instead of providing a code string like "ccDdddddddddd", we could have been more explicit about the specific columns for each type.
read_csv('noaa/USC00200230.csv',
col_types=cols(
STATION = col_character(),
NAME = col_character(),
DATE = col_date(),
DAPR = col_double(),
DASF = col_double(),
MDPR = col_double(),
MDSF = col_double(),
PRCP = col_double(),
SNOW = col_double(),
SNWD = col_double(),
TMAX = col_double(),
TMIN = col_double(),
TOBS = col_double())
)
## # A tibble: 46,650 x 13
## STATION NAME DATE DAPR DASF MDPR MDSF PRCP SNOW SNWD TMAX
## <chr> <chr> <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 USC002… ANN … 1891-10-01 NA NA NA NA NA NA NA 20.6
## 2 USC002… ANN … 1891-10-02 NA NA NA NA NA NA NA 26.7
## 3 USC002… ANN … 1891-10-03 NA NA NA NA NA NA NA 26.1
## 4 USC002… ANN … 1891-10-04 NA NA NA NA 7.6 NA NA 22.8
## 5 USC002… ANN … 1891-10-05 NA NA NA NA NA NA NA 13.9
## 6 USC002… ANN … 1891-10-06 NA NA NA NA NA NA NA 14.4
## 7 USC002… ANN … 1891-10-07 NA NA NA NA 4.1 NA NA 10.6
## 8 USC002… ANN … 1891-10-08 NA NA NA NA NA NA NA 13.9
## 9 USC002… ANN … 1891-10-09 NA NA NA NA NA NA NA 15
## 10 USC002… ANN … 1891-10-10 NA NA NA NA NA NA NA 16.7
## # … with 46,640 more rows, and 2 more variables: TMIN <dbl>, TOBS <dbl>
As we saw earlier, there’s also col_logical. Since so many of our columns are of type col_double, we can make that the default setting and simplify the syntax
read_csv('noaa/USC00200230.csv',
col_types=cols(
STATION = col_character(),
NAME = col_character(),
DATE = col_date(),
.default = col_double())
)
## # A tibble: 46,650 x 13
## STATION NAME DATE DAPR DASF MDPR MDSF PRCP SNOW SNWD TMAX
## <chr> <chr> <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 USC002… ANN … 1891-10-01 NA NA NA NA NA NA NA 20.6
## 2 USC002… ANN … 1891-10-02 NA NA NA NA NA NA NA 26.7
## 3 USC002… ANN … 1891-10-03 NA NA NA NA NA NA NA 26.1
## 4 USC002… ANN … 1891-10-04 NA NA NA NA 7.6 NA NA 22.8
## 5 USC002… ANN … 1891-10-05 NA NA NA NA NA NA NA 13.9
## 6 USC002… ANN … 1891-10-06 NA NA NA NA NA NA NA 14.4
## 7 USC002… ANN … 1891-10-07 NA NA NA NA 4.1 NA NA 10.6
## 8 USC002… ANN … 1891-10-08 NA NA NA NA NA NA NA 13.9
## 9 USC002… ANN … 1891-10-09 NA NA NA NA NA NA NA 15
## 10 USC002… ANN … 1891-10-10 NA NA NA NA NA NA NA 16.7
## # … with 46,640 more rows, and 2 more variables: TMIN <dbl>, TOBS <dbl>
All that to read in the data without an error :) Hopefully, you can appreciate the flexibility that read_csv gives you for specifying the type of data that you are reading in.
selecting-ing columns from a data frame
Using our skills that we developed with the Project Tycho data we can begin to take a look at the structure of these data. We can see that there are 46,642 rows (46,642 days / 365 days/year ~ 128 years worth of data) and 12 columns. Some of these columns aren’t very useful (e.g. STATION and NAME are the same value for every row) and some of them don’t have much data (e.g. DAPR, DASF, MDPR, MDSF have 0% coverage). We can either remove those columns or we can select the columns we want to keep. To do either approach we can use the select function.
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(DATE)
## # A tibble: 46,650 x 1
## DATE
## <date>
## 1 1891-10-01
## 2 1891-10-02
## 3 1891-10-03
## 4 1891-10-04
## 5 1891-10-05
## 6 1891-10-06
## 7 1891-10-07
## 8 1891-10-08
## 9 1891-10-09
## 10 1891-10-10
## # … with 46,640 more rows
This returns a new data frame that is made up of the data in the DATE column. We want more columns. We can add column names separated by commas
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(DATE, TMAX)
## # A tibble: 46,650 x 2
## DATE TMAX
## <date> <dbl>
## 1 1891-10-01 20.6
## 2 1891-10-02 26.7
## 3 1891-10-03 26.1
## 4 1891-10-04 22.8
## 5 1891-10-05 13.9
## 6 1891-10-06 14.4
## 7 1891-10-07 10.6
## 8 1891-10-08 13.9
## 9 1891-10-09 15
## 10 1891-10-10 16.7
## # … with 46,640 more rows
Let’s get all of the temperature data
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(DATE, TMAX, TMIN, TOBS)
## # A tibble: 46,650 x 4
## DATE TMAX TMIN TOBS
## <date> <dbl> <dbl> <dbl>
## 1 1891-10-01 20.6 7.8 NA
## 2 1891-10-02 26.7 13.9 NA
## 3 1891-10-03 26.1 16.1 NA
## 4 1891-10-04 22.8 11.1 NA
## 5 1891-10-05 13.9 6.7 NA
## 6 1891-10-06 14.4 5 NA
## 7 1891-10-07 10.6 5.6 NA
## 8 1891-10-08 13.9 3.3 NA
## 9 1891-10-09 15 3.9 NA
## 10 1891-10-10 16.7 5 NA
## # … with 46,640 more rows
Three column names for temperature isn’t too horrible to type in. There’s an easier way that you might find useful in other circumstances. We can select columns that starts_with a “T”
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(DATE, starts_with("T"))
## # A tibble: 46,650 x 4
## DATE TMAX TMIN TOBS
## <date> <dbl> <dbl> <dbl>
## 1 1891-10-01 20.6 7.8 NA
## 2 1891-10-02 26.7 13.9 NA
## 3 1891-10-03 26.1 16.1 NA
## 4 1891-10-04 22.8 11.1 NA
## 5 1891-10-05 13.9 6.7 NA
## 6 1891-10-06 14.4 5 NA
## 7 1891-10-07 10.6 5.6 NA
## 8 1891-10-08 13.9 3.3 NA
## 9 1891-10-09 15 3.9 NA
## 10 1891-10-10 16.7 5 NA
## # … with 46,640 more rows
Other helper functions like starts_with that you can use with select include ends_with, contains, and matches. Let’s put this together with the precipitation data that we might be interested in using later
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(DATE, starts_with("T"), PRCP, SNOW, SNWD)
## # A tibble: 46,650 x 7
## DATE TMAX TMIN TOBS PRCP SNOW SNWD
## <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1891-10-01 20.6 7.8 NA NA NA NA
## 2 1891-10-02 26.7 13.9 NA NA NA NA
## 3 1891-10-03 26.1 16.1 NA NA NA NA
## 4 1891-10-04 22.8 11.1 NA 7.6 NA NA
## 5 1891-10-05 13.9 6.7 NA NA NA NA
## 6 1891-10-06 14.4 5 NA NA NA NA
## 7 1891-10-07 10.6 5.6 NA 4.1 NA NA
## 8 1891-10-08 13.9 3.3 NA NA NA NA
## 9 1891-10-09 15 3.9 NA NA NA NA
## 10 1891-10-10 16.7 5 NA NA NA NA
## # … with 46,640 more rows
We can also tell select, which columns we don’t want. This can be done by putting a - before the column name. For instance, if we wanted to remove the STATION and NAME columns we could do this
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(-STATION, -NAME)
## # A tibble: 46,650 x 11
## DATE DAPR DASF MDPR MDSF PRCP SNOW SNWD TMAX TMIN TOBS
## <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1891-10-01 NA NA NA NA NA NA NA 20.6 7.8 NA
## 2 1891-10-02 NA NA NA NA NA NA NA 26.7 13.9 NA
## 3 1891-10-03 NA NA NA NA NA NA NA 26.1 16.1 NA
## 4 1891-10-04 NA NA NA NA 7.6 NA NA 22.8 11.1 NA
## 5 1891-10-05 NA NA NA NA NA NA NA 13.9 6.7 NA
## 6 1891-10-06 NA NA NA NA NA NA NA 14.4 5 NA
## 7 1891-10-07 NA NA NA NA 4.1 NA NA 10.6 5.6 NA
## 8 1891-10-08 NA NA NA NA NA NA NA 13.9 3.3 NA
## 9 1891-10-09 NA NA NA NA NA NA NA 15 3.9 NA
## 10 1891-10-10 NA NA NA NA NA NA NA 16.7 5 NA
## # … with 46,640 more rows
I usually only use this approach if there are one or two columns that I want to remove.
Let’s return to the earlier way we set up the select function…
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(DATE, starts_with("T"), PRCP, SNOW, SNWD)
## # A tibble: 46,650 x 7
## DATE TMAX TMIN TOBS PRCP SNOW SNWD
## <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1891-10-01 20.6 7.8 NA NA NA NA
## 2 1891-10-02 26.7 13.9 NA NA NA NA
## 3 1891-10-03 26.1 16.1 NA NA NA NA
## 4 1891-10-04 22.8 11.1 NA 7.6 NA NA
## 5 1891-10-05 13.9 6.7 NA NA NA NA
## 6 1891-10-06 14.4 5 NA NA NA NA
## 7 1891-10-07 10.6 5.6 NA 4.1 NA NA
## 8 1891-10-08 13.9 3.3 NA NA NA NA
## 9 1891-10-09 15 3.9 NA NA NA NA
## 10 1891-10-10 16.7 5 NA NA NA NA
## # … with 46,640 more rows
Renaming columns in a data frame
I’m not a big fan of these column names. I have a few “rules” for column names…
- Names should be descriptive (e.g.
date, notx) - No spaces in names, use underscores (e.g.
t_maxnott max) - Keep cases consistent; preference for all lower case (e.g.
t_maxandt_min, notT_MaxandT_min)
The idea is that the column names should be easy for our computers to read and easy for us to read. SNWD is easy for our computer to read, but I’m not really sure that I’ll remember “snow depth in millimeters” when I come back to this dataset in a month. Let’s give these columns names that are a bit more meaningful. We can do this with the rename function. The syntax of this function is
data %>%
rename("new_name" = "old_name")
You don’t always need the quote marks around the column names, but to be consistent, I always use them. Applying to our data we can do the following
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(DATE, starts_with("T"), PRCP, SNOW, SNWD) %>%
rename("date" = "DATE")
## # A tibble: 46,650 x 7
## date TMAX TMIN TOBS PRCP SNOW SNWD
## <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1891-10-01 20.6 7.8 NA NA NA NA
## 2 1891-10-02 26.7 13.9 NA NA NA NA
## 3 1891-10-03 26.1 16.1 NA NA NA NA
## 4 1891-10-04 22.8 11.1 NA 7.6 NA NA
## 5 1891-10-05 13.9 6.7 NA NA NA NA
## 6 1891-10-06 14.4 5 NA NA NA NA
## 7 1891-10-07 10.6 5.6 NA 4.1 NA NA
## 8 1891-10-08 13.9 3.3 NA NA NA NA
## 9 1891-10-09 15 3.9 NA NA NA NA
## 10 1891-10-10 16.7 5 NA NA NA NA
## # … with 46,640 more rows
We can add other column name changes to the rename function by separating them with a comma
read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(DATE, starts_with("T"), PRCP, SNOW, SNWD) %>%
rename("date" = "DATE", "t_max_c" = "TMAX", "t_min_c" = "TMIN", "t_obs_c" = "TOBS", "total_precip_mm" = "PRCP", "snow_fall_mm" = "SNOW", "snow_depth_mm" = "SNWD")
## # A tibble: 46,650 x 7
## date t_max_c t_min_c t_obs_c total_precip_mm snow_fall_mm snow_depth_mm
## <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1891-10-01 20.6 7.8 NA NA NA NA
## 2 1891-10-02 26.7 13.9 NA NA NA NA
## 3 1891-10-03 26.1 16.1 NA NA NA NA
## 4 1891-10-04 22.8 11.1 NA 7.6 NA NA
## 5 1891-10-05 13.9 6.7 NA NA NA NA
## 6 1891-10-06 14.4 5 NA NA NA NA
## 7 1891-10-07 10.6 5.6 NA 4.1 NA NA
## 8 1891-10-08 13.9 3.3 NA NA NA NA
## 9 1891-10-09 15 3.9 NA NA NA NA
## 10 1891-10-10 16.7 5 NA NA NA NA
## # … with 46,640 more rows
Nice, eh? One other stylistic comment is that the arguments for our rename function are quite long and not super easy to read. We can make the code more readable by putting a line beak in after commas and we can use a tab to indicate that the arguments go with the function like this. Let’s also assign the output of this pipeline to the variable aa_weather
aa_weather <- read_csv('noaa/USC00200230.csv', col_types="ccDdddddddddd") %>%
select(DATE, starts_with("T"), PRCP, SNOW, SNWD) %>%
rename("date" = "DATE",
"t_max_c" = "TMAX",
"t_min_c" = "TMIN",
"t_obs_c" = "TOBS",
"total_precip_mm" = "PRCP",
"snow_fall_mm" = "SNOW",
"snow_depth_mm" = "SNWD")
We’ve done quite a bit here without really “doing anything”. But we have! We’ve made the data easier to work with.
Exercises
1. What are the strengths and weaknesses of the following variable/column names?
is_cumulative_countxheightfootemp_clongcolor
2. Revisit the Project Tycho data that we worked with earlier. Can you write a pipeline consisting of read_csv, select, and rename to create a data frame that includes the period’s end date, state, counts, and whether the counts are cumulative? Be sure to give meaningful column names and tell read_csv, the data type in each column
3. Can you find a weather station with a long history of data from your favorite location?