Session 14
Topics
- Thinking through questions
- Finding text with R
- Adjusting the shape of a data frame
- More sophisticated text searches
- Modifying text based on patterns
Thinking through questions
In the last several sessions, we looked at gender and racial/ethnic representation at pretty granular levels - the level of institution type and particular institutions. I’d like to get a bit more refined and look within a specific set of disciplines. I’m going to start the analysis with “Microbiology”. To do this analysis, I need to develop a sense of how the data are reported. A “microbiologist” is a very diverse concept! They can study organisms in any of the three domains of life, viruses, interactions with animals or plants or the natural environment. They can be engineered, host-associated, or natural ecosystems. They can be studied at the gene, protein, organismal, or community level. Depending on the campus, the people that study microbes at these various levels can be one person in a Biology department or 20 in a Microbiology department or 200 across 20 different departments. How we organize data into a framework is critical to the interpretation of our data. We could be the best R programmer in the world, but if we can’t get the definitions right, we’re toast.
Inspecting the CIP codes in ipeds/c2018_a.xlsx, I see that there’s some organization to them - the first two numbers correspond to a broad discipline (e.g. 26 - Biology or 27 - Math). Naturally, I immediately noticed that “26.1102 - Biostatistics” is listed under Biology and not under Math and “27.0306 - Mathematical Biology” is listed under Math rather than Biology. For the microbiology-affiliated fields I listed above, I notice that their CIP codes all start with “26.05”. Among those CIP codes, there are
- 26.0502 Microbiology, General
- 26.0503 Medical Microbiology and Bacteriology
- 26.0504 Virology
- 26.0505 Parasitology
- 26.0507 Immunology
- 26.0508 Microbiology and Immunology
- 26.0599 Microbiological Sciences and Immunology, Other
Of course, there are Immunologists that don’t think much about microbes and there are card carrying microbiologists in general Biology departments and even Engineering. This overlap between subdisciplines is why it is a bad idea to aggregate all institutions blindly. A better practice would be to pick peer institutions where one has a better understanding of where a microbiologist would most likely be trained within that institution. In other words, take this analysis with a grain of salt. We’re here to learn R, not make grand statements about bias in science.
Finding text with R
This discussion gets us pretty close to figuring out the categories we’re interested in, but let’s step back and see if we can do a more systematic review of where microbiology shows up in the CIP codes.
library(tidyverse)
library(readxl)
cip_codes <- read_excel("ipeds/c2018_a.xlsx", sheet="Frequencies") %>%
filter(varname == "CIPCODE" & codevalue != "99") %>%
select(codevalue, valuelabel) %>%
rename(cip_code = codevalue,
description = valuelabel)
As I pointed out above, it seems pretty clear that microbiology disciplines are “supposed” to have a CIP code that starts “26.05”. I’m curious whether “Microbiology” shows up in any other titles. To figure this out, we need to learn a new function - str_detect. This function tells us whether a character variable contains a string we are interested in. For example, consider this character vector
baseball_teams <- c("cubs", "white sox", "red sox", "cardinals", "yankees")
I want to know which values in baseball_teams has the word “sox”. I can use str_detect
str_detect(baseball_teams, "sox")
## [1] FALSE TRUE TRUE FALSE FALSE
If I wanted to know which elements in baseball_teams had “sox”, I could use str_which
str_which(baseball_teams, "sox")
## [1] 2 3
And if I wanted to get the values that contained “sox”, I would use str_subset….
str_subset(baseball_teams, "sox")
## [1] "white sox" "red sox"
The nice thing about str_detect is that because it returns a logical value, I can use it with filter to get those rows from a data frame that match the search string
cip_codes %>%
filter(str_detect(cip_code, "26.05"))
## # A tibble: 7 x 2
## cip_code description
## <chr> <chr>
## 1 26.0502 Microbiology, General
## 2 26.0503 Medical Microbiology and Bacteriology
## 3 26.0504 Virology
## 4 26.0505 Parasitology
## 5 26.0507 Immunology
## 6 26.0508 Microbiology and Immunology
## 7 26.0599 Microbiological Sciences and Immunology, Other
Let’s look for microbiology elsewhere by using str_detect on the description column
cip_codes %>%
filter(str_detect(description, "Microb"))
## # A tibble: 5 x 2
## cip_code description
## <chr> <chr>
## 1 26.0502 Microbiology, General
## 2 26.0503 Medical Microbiology and Bacteriology
## 3 26.0508 Microbiology and Immunology
## 4 26.0599 Microbiological Sciences and Immunology, Other
## 5 51.2504 Veterinary Microbiology and Immunobiology
Aha, we found a “Veterinary Microbiology and Immunology”.
Let’s look a bit broader for those descriptions that have “biology” in them
cip_codes %>%
filter(str_detect(description, "biology"))
## # A tibble: 10 x 2
## cip_code description
## <chr> <chr>
## 1 26.0209 Radiation Biology/Radiobiology
## 2 26.0502 Microbiology, General
## 3 26.0503 Medical Microbiology and Bacteriology
## 4 26.0508 Microbiology and Immunology
## 5 26.1503 Neurobiology and Anatomy
## 6 26.1504 Neurobiology and Behavior
## 7 26.1599 Neurobiology and Neurosciences, Other
## 8 42.2706 Physiological Psychology/Psychobiology
## 9 51.2504 Veterinary Microbiology and Immunobiology
## 10 51.2505 Veterinary Pathology and Pathobiology
of course, we could have also searched for
cip_codes %>%
filter(str_detect(description, "Biology"))
## # A tibble: 36 x 2
## cip_code description
## <chr> <chr>
## 1 03.0502 Forest Sciences and Biology
## 2 13.1322 Biology Teacher Education
## 3 26.0101 Biology/Biological Sciences, General
## 4 26.0204 Molecular Biology
## 5 26.0207 Structural Biology
## 6 26.0209 Radiation Biology/Radiobiology
## 7 26.0210 Biochemistry and Molecular Biology
## 8 26.0299 Biochemistry, Biophysics and Molecular Biology, Other
## 9 26.0301 Botany/Plant Biology
## 10 26.0308 Plant Molecular Biology
## # … with 26 more rows
or
cip_codes %>%
filter(str_detect(description, "Biolog"))
## # A tibble: 41 x 2
## cip_code description
## <chr> <chr>
## 1 03.0502 Forest Sciences and Biology
## 2 13.1322 Biology Teacher Education
## 3 14.4501 Biological/Biosystems Engineering
## 4 26.0101 Biology/Biological Sciences, General
## 5 26.0204 Molecular Biology
## 6 26.0207 Structural Biology
## 7 26.0209 Radiation Biology/Radiobiology
## 8 26.0210 Biochemistry and Molecular Biology
## 9 26.0299 Biochemistry, Biophysics and Molecular Biology, Other
## 10 26.0301 Botany/Plant Biology
## # … with 31 more rows
Developing a meaningful search term is a challenge. If we are too broad (e.g. “Micro”) we could get something that isn’t relevant like “Data Entry/Microcomputer Applications”. If we’re too specific (e.g. “Biological” vs. “Biology” vs “biology”), then we risk losing out on relevant matches. To make our search insensitive to the case of the search string
cip_codes %>%
filter(str_detect(description, "(?i)Biolog"))
## # A tibble: 51 x 2
## cip_code description
## <chr> <chr>
## 1 03.0502 Forest Sciences and Biology
## 2 13.1322 Biology Teacher Education
## 3 14.4501 Biological/Biosystems Engineering
## 4 26.0101 Biology/Biological Sciences, General
## 5 26.0204 Molecular Biology
## 6 26.0207 Structural Biology
## 7 26.0209 Radiation Biology/Radiobiology
## 8 26.0210 Biochemistry and Molecular Biology
## 9 26.0299 Biochemistry, Biophysics and Molecular Biology, Other
## 10 26.0301 Botany/Plant Biology
## # … with 41 more rows
Notice that this will match both “Biology/Biological Sciences, General” and “Microbiology, General”. The “(?i)Biolog” is what is called a “regular expression”. Believe it or not, this is a relatively simple regular expression. We can use far more complicated patterns to identify dates, email addresses, genes, and more.
If it wasn’t already clear, defining “biology” or “microbiology” is pretty hard! Let’s stick with the “26.05” CIP code stub to define microbiology
microbiology_cips <- read_excel("ipeds/c2018_a.xlsx", sheet="Frequencies") %>%
filter(varname == "CIPCODE" & codevalue != "99") %>%
select(codevalue, valuelabel) %>%
rename(cip_code = codevalue,
description = valuelabel) %>%
filter(str_detect(cip_code, "26.05"))
Which institutions have enough microbiology on campus that they feel comfortable using one of these codes? To answer this question, I could now do an inner join between microbiology and the data in ipeds/c2018_a.csv to find the UNITID values that reported graduates in one of these CIP codes. Then I could join those results with the broader institutional data in ipeds/hd2018.csv. All of this should sound familiar by now.
microbiology_degrees <- read_csv("ipeds/c2018_a.csv") %>%
inner_join(microbiology_cips, ., by=c("cip_code" = "CIPCODE")) %>%
filter(MAJORNUM == 1 & CTOTALT > 0 & (AWLEVEL == "05" | AWLEVEL == "17"))
Adjusting the shape of a data frame
Before we go all the way, let’s see how many institutions claimed each type of microbiology for each degree type.
microbiology_degrees %>%
count(description, AWLEVEL)
## # A tibble: 10 x 3
## description AWLEVEL n
## <chr> <chr> <int>
## 1 Immunology 17 28
## 2 Medical Microbiology and Bacteriology 05 11
## 3 Medical Microbiology and Bacteriology 17 26
## 4 Microbiological Sciences and Immunology, Other 05 1
## 5 Microbiological Sciences and Immunology, Other 17 11
## 6 Microbiology and Immunology 05 7
## 7 Microbiology and Immunology 17 17
## 8 Microbiology, General 05 77
## 9 Microbiology, General 17 47
## 10 Virology 17 3
That’s a little hard for me to read, what if we make the column names the different degree types? Remember how to do that?
microbiology_degrees %>%
count(description, AWLEVEL) %>%
pivot_wider(names_from=AWLEVEL, values_from=n, values_fill=list(n=0)) %>% # if we leave out `values_fill we get NA values`
select(description, `05`, `17`)
## # A tibble: 6 x 3
## description `05` `17`
## <chr> <int> <int>
## 1 Immunology 0 28
## 2 Medical Microbiology and Bacteriology 11 26
## 3 Microbiological Sciences and Immunology, Other 1 11
## 4 Microbiology and Immunology 7 17
## 5 Microbiology, General 77 47
## 6 Virology 0 3
From this, it looks like most bachelor’s degrees in Microbiology are in “Microbiology, General” and occasionally in “Medical Microbiology and Bacteriology”. On the other hand, there is more specialization at the doctorate level. Back to our question of which schools are these… I will modify microbiology_degrees to get the total number of graduates with a bachelor’s and doctorate degree from each major as separate columns
microbiology_degrees %>%
select(UNITID, description, AWLEVEL, CTOTALT) %>%
pivot_wider(names_from=AWLEVEL, values_from=CTOTALT, values_fill=list(n=0))
## # A tibble: 196 x 4
## UNITID description `17` `05`
## <dbl> <chr> <dbl> <dbl>
## 1 100663 Microbiology, General 13 NA
## 2 100724 Microbiology, General 2 NA
## 3 100751 Microbiology, General NA 15
## 4 104151 Microbiology, General 2 65
## 5 104179 Microbiology, General 1 44
## 6 105330 Microbiology, General NA 27
## 7 110404 Microbiology, General 1 NA
## 8 110422 Microbiology, General NA 10
## 9 110538 Microbiology, General NA 24
## 10 110583 Microbiology, General NA 21
## # … with 186 more rows
This code chunk shows there are 196 institution/department combinations that awarded a microbiology degree in 2018. Are there institutions that show up multiple times?
microbiology_degrees %>%
select(UNITID, description, AWLEVEL, CTOTALT) %>%
pivot_wider(names_from=AWLEVEL, values_from=CTOTALT, values_fill=list(n=0)) %>%
count(UNITID) %>%
count(n)
## # A tibble: 4 x 2
## n nn
## <int> <int>
## 1 1 139
## 2 2 22
## 3 3 3
## 4 4 1
It appears that 139 schools offer one microbiology major, 22 offer two, 3 offer three, and 1 offers four. How many does the University of Michigan offer?
microbiology_degrees %>%
select(UNITID, description, AWLEVEL, CTOTALT) %>%
pivot_wider(names_from=AWLEVEL, values_from=CTOTALT, values_fill=list(n=0)) %>%
count(UNITID) %>%
filter(UNITID == 170976)
## # A tibble: 1 x 2
## UNITID n
## <dbl> <int>
## 1 170976 3
3!?! I know there’s “Microbiology and Immunology”, “Immunology” and… I’m not sure. Let’s find out.
microbiology_degrees %>%
select(UNITID, description, AWLEVEL, CTOTALT) %>%
pivot_wider(names_from=AWLEVEL, values_from=CTOTALT, values_fill=list(n=0)) %>%
filter(UNITID == 170976)
## # A tibble: 3 x 4
## UNITID description `17` `05`
## <dbl> <chr> <dbl> <dbl>
## 1 170976 Microbiology, General NA 56
## 2 170976 Immunology 3 NA
## 3 170976 Microbiology and Immunology 9 NA
Ah, yes. The undergrads - “Microbiology, General”. Instead of looking up UNITID codes manually, let’s bring those in with a join
micro_degree_counts <- microbiology_degrees %>%
select(UNITID, description, AWLEVEL, CTOTALT) %>%
pivot_wider(names_from=AWLEVEL, values_from=CTOTALT, values_fill=list(n=0)) %>%
inner_join(read_csv("ipeds/hd2018.csv"), ., by="UNITID") %>%
select(UNITID, INSTNM, description, `17`, `05`)
Now it’s easier to see how many different microbiology majors these schools offer
micro_degree_counts %>%
count(INSTNM) %>%
arrange(desc(n))
## # A tibble: 165 x 2
## INSTNM n
## <chr> <int>
## 1 University of Pittsburgh-Pittsburgh Campus 4
## 2 Harvard University 3
## 3 University of Michigan-Ann Arbor 3
## 4 Washington University in St Louis 3
## 5 Baylor College of Medicine 2
## 6 Boston University 2
## 7 Cornell University 2
## 8 Iowa State University 2
## 9 North Carolina State University at Raleigh 2
## 10 Pennsylvania State University-Main Campus 2
## # … with 155 more rows
More sophisticated text searches
Looking through these institutions, it’s pretty clear that to have a dedicated microbiology program of any type suggests that the institution is research focused. We can also see that the program to graduate the most doctorates, University of Wisconsin-Madison, only awarded 19 PhDs in 2018. Based on this, it probably doesn’t make much sense to compare the representation at individual institutions for a single year. Now that we have a better handle on the data, let’s see what the gender representation looks like for microbiologists awarded bachelor’s and doctorate degrees in 2018.
microbiology_degrees %>%
select(UNITID, description, AWLEVEL, CTOTALW, CTOTALT) %>%
group_by(AWLEVEL) %>%
summarize(f_women = sum(CTOTALW)/sum(CTOTALT))
## # A tibble: 2 x 2
## AWLEVEL f_women
## <chr> <dbl>
## 1 05 0.580
## 2 17 0.571
The fraction of women receiving a bachelors degree and a PhD are fairly similar in microbiology. What does this fraction look like for all of the biology CIP codes? To answer this, I can modify the code chunk from above where we defined microbiology_cips
biology_cips <- read_excel("ipeds/c2018_a.xlsx", sheet="Frequencies") %>%
filter(varname == "CIPCODE" & codevalue != "99") %>%
select(codevalue, valuelabel) %>%
rename(cip_code = codevalue,
description = valuelabel) %>%
filter(str_detect(cip_code, "26"))
If you look at biology_cips you’ll notice a small problem. We identified any CIP code that contained “26”, which got us things like “Italian Studies” and “German Language Teacher Education”. Oops. We what CIP codes that start with “26”. We can modify our search term to use an “anchor”. The ^ tells str_detect to look at the beginning of a string and $ tells it to look at the end of the string.
biology_cips <- read_excel("ipeds/c2018_a.xlsx", sheet="Frequencies") %>%
filter(varname == "CIPCODE" & codevalue != "99") %>%
select(codevalue, valuelabel) %>%
rename(cip_code = codevalue,
description = valuelabel) %>%
filter(str_detect(cip_code, "^26"))
That’s more like it. We can create biology_degrees similar to how we generated microbiology_degrees
biology_degrees <- read_csv("ipeds/c2018_a.csv") %>%
inner_join(biology_cips, ., by=c("cip_code" = "CIPCODE")) %>%
filter(MAJORNUM == 1 & CTOTALT > 0 & (AWLEVEL == "05" | AWLEVEL == "17"))
And we can calculate the fraction of women across all biology for both degree levels
biology_degrees %>%
select(UNITID, description, AWLEVEL, CTOTALW, CTOTALT) %>%
group_by(AWLEVEL) %>%
summarize(f_women = sum(CTOTALW)/sum(CTOTALT))
## # A tibble: 2 x 2
## AWLEVEL f_women
## <chr> <dbl>
## 1 05 0.622
## 2 17 0.533
Interesting, it appears that in some fields there is a larger drop in representation of women. The individual number of graduates in each sub discipline are too small to tell us much. However, if we aggregated the data at the “hundredths” place in the CIP code we could look at a broader level that is still more resolved than all of biology.
Modifying text based on patterns
How do we extract “26.05” from “26.0502”? How do we do extract “26.01” from “26.0101” with the same command? We need another function that works with strings - str_replace. This function takes a set of strings, a pattern to match, and a replacement value. Here’s the brute force way to do it for one CIP code
cip_code <- "26.0502"
str_replace(cip_code, pattern="26.0502", replacement="26.05")
## [1] "26.05"
The challenge is that we can’t write hundreds of str_replace functions. Also, if we reanalyze the data when the 2019 data are released, we may need to add more str_replace function calls. No fun. We need to learn a little more about regular expressions. We can use “metacharacters” to represent a generic letter, number, or space. Here’s a quick set of examples…
x <- "My phone number is (555)678-1234"
#str_replace only replaces the first matched value
str_replace_all(x, "\\w", "x") # match any alphanumeric character
## [1] "xx xxxxx xxxxxx xx (xxx)xxx-xxxx"
str_replace_all(x, "\\d", "#") # match any number
## [1] "My phone number is (###)###-####"
str_replace_all(x, "\\s", "_") # match any space
## [1] "My_phone_number_is_(555)678-1234"
str_replace_all(x, "[()-]", "_") # match any (, ), or -
## [1] "My phone number is _555_678_1234"
We can match more than a single character
str_replace_all(x, "\\d\\d\\d\\d", "####") # match any number
## [1] "My phone number is (555)678-####"
What does that mean for our CIP code?
str_replace(cip_code, "\\d\\d$", "")
## [1] "26.05"
Let’s apply this to group by the sub fields within biology and calculate the percent drop going from bachelor’s to doctorate degrees
biology_degrees %>%
select(UNITID, cip_code, description, AWLEVEL, CTOTALW, CTOTALT) %>%
mutate(sub_category = str_replace(cip_code, "\\d\\d$", "")) %>%
group_by(sub_category, AWLEVEL) %>%
summarize(f_women = sum(CTOTALW)/sum(CTOTALT)) %>%
ungroup() %>%
pivot_wider(names_from=AWLEVEL, values_from=f_women) %>%
mutate(percent_drop = 100*(`05`-`17`)/`05`) %>%
arrange(percent_drop)
## # A tibble: 15 x 4
## sub_category `05` `17` percent_drop
## <chr> <dbl> <dbl> <dbl>
## 1 26.05 0.580 0.571 1.52
## 2 26.13 0.643 0.612 4.70
## 3 26.10 0.584 0.554 5.20
## 4 26.02 0.521 0.486 6.81
## 5 26.11 0.480 0.437 8.87
## 6 26.04 0.580 0.521 10.0
## 7 26.03 0.536 0.471 12.2
## 8 26.99 0.690 0.603 12.6
## 9 26.15 0.655 0.563 14.0
## 10 26.09 0.588 0.5 15.0
## 11 26.01 0.637 0.530 16.9
## 12 26.08 0.705 0.578 18.1
## 13 26.12 0.536 0.4 25.4
## 14 26.07 0.688 0.502 27.1
## 15 26.14 NA 0.528 NA
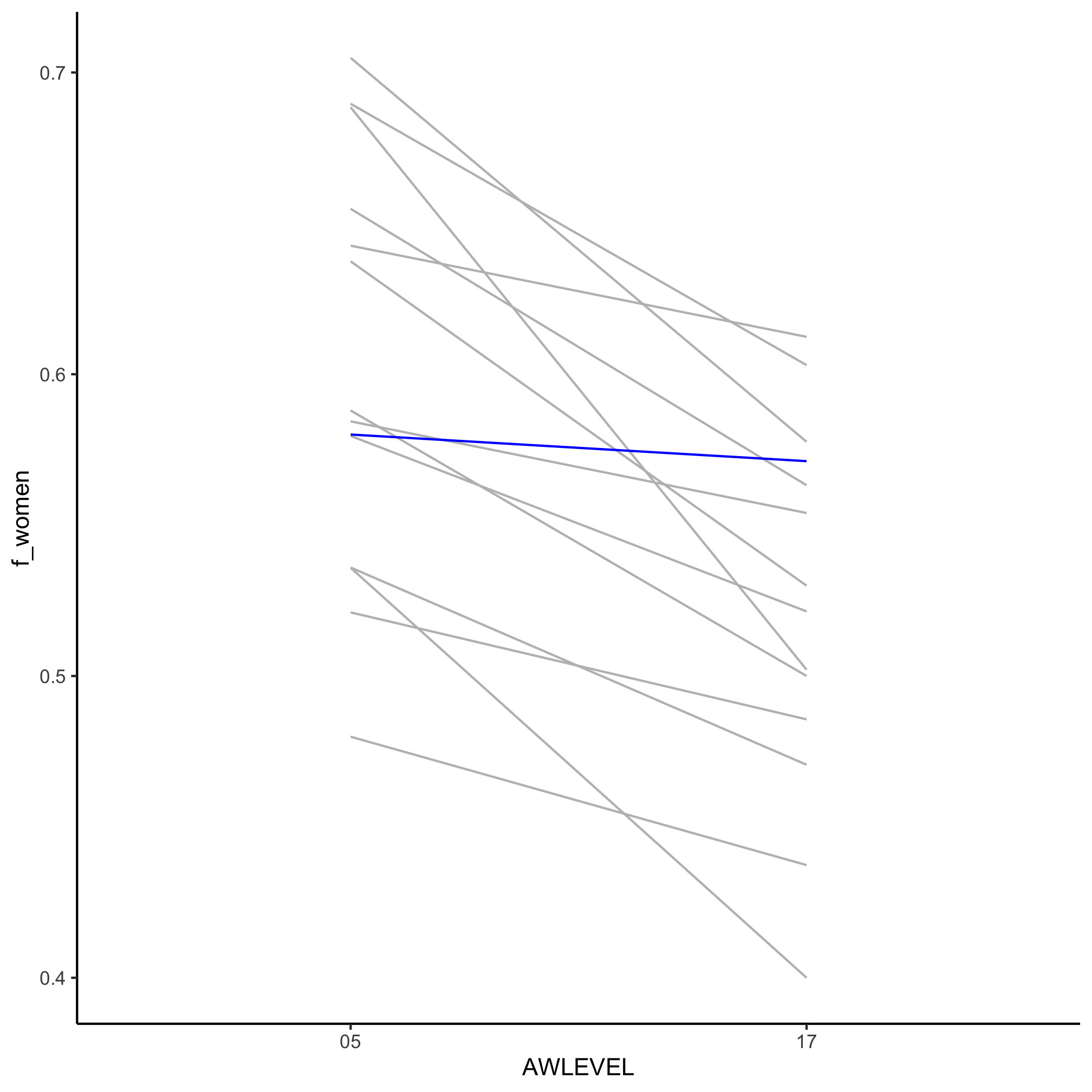
It appears that of all the biology disciplines, microbiology does the best job of retaining its gender balance between the bachelor’s and doctorate degree awardees.
Exercises
1. It is interesting that after microbiology, sub field “26.13” is next in the ranking for retaining women and has more than 60% of its degrees going to women at both levels. What subfields are in that grouping? The worst at retention is “26.07”. Which field is that?
2. Generate a slopegraph of the biology CIP codes with each line representing a different sub field (i.e. to the level of XX.XX). Can you highlight the microbiology line?
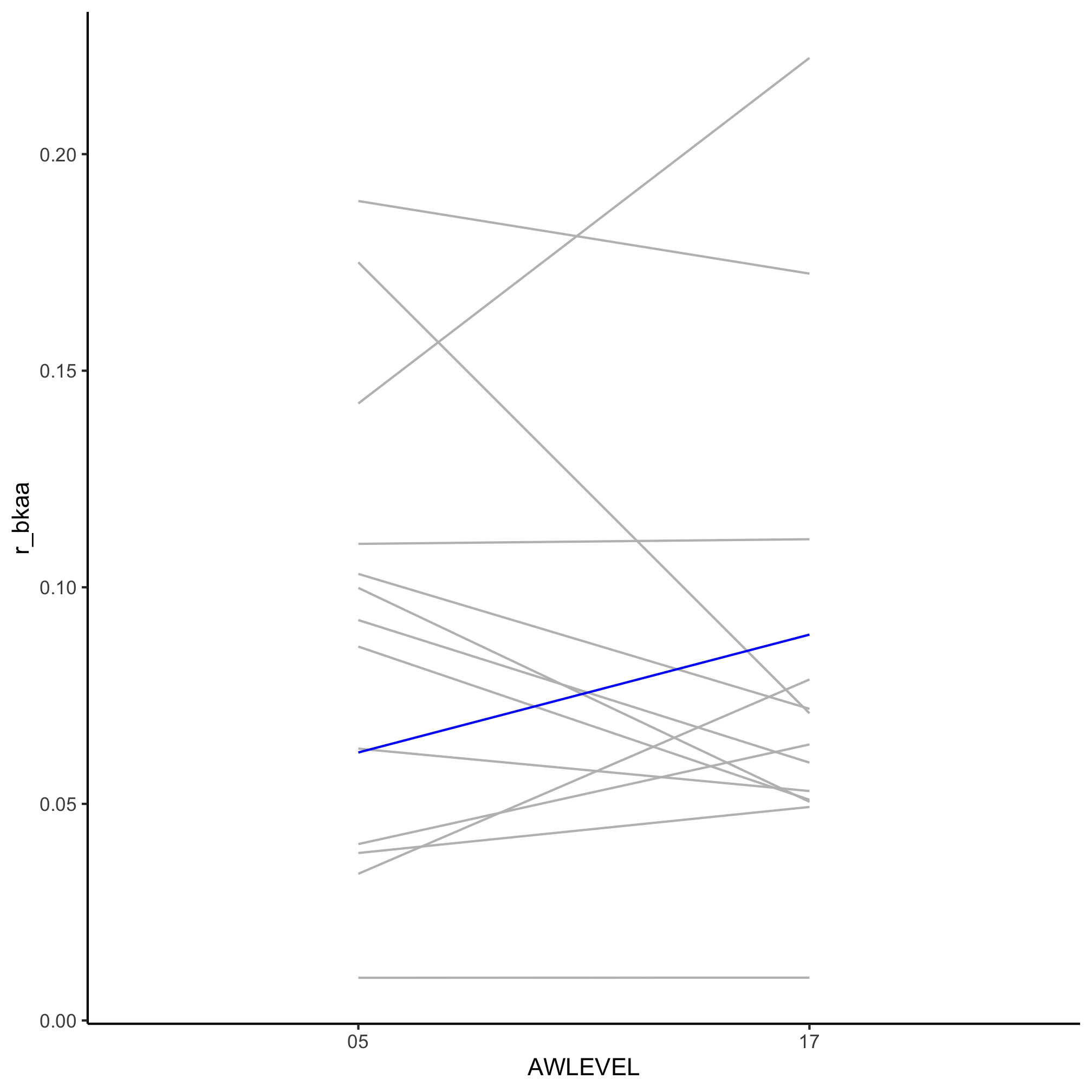
3. Can you repeat the analysis resulting in the slope graph in the last exercise, but looking at the representation of “Black and African American” awardees as a ratio of “White” awardees?
4. Of the engineering sub fields with more than 100 bachelor’s and 100 doctorate degrees, which has the best retention and representation of women? The worst?