Session 4: Combining and Exploring Data Frames
Topics
- Merging data frames
- Selecting columns from data frames
- Selecting rows from data frames
- Connecting steps in data processing with pipes
- Boolean operations
Joining metadata and pcoa
If we compile all of the code in the last lesson, we now have this code chunk which is a more sophisticated version of the original code chunk that we created in the first lesson.
library(tidyverse)
library(readxl)
pcoa <- read_tsv(file="raw_data/baxter.braycurtis.pcoa.axes",
col_types=cols(group=col_character())
)
metadata <- read_excel(path="raw_data/baxter.metadata.xlsx",
col_types=c(sample = "text", fit_result = "numeric", Site = "text", Dx_Bin = "text",
dx = "text", Hx_Prev = "logical", Hx_of_Polyps = "logical", Age = "numeric",
Gender = "text", Smoke = "logical", Diabetic = "logical", Hx_Fam_CRC = "logical",
Height = "numeric", Weight = "numeric", NSAID = "logical", Diabetes_Med = "logical",
stage = "text")
)
metadata <- mutate(metadata, Height = na_if(Height, 0))
metadata <- mutate(metadata, Weight = na_if(Weight, 0))
metadata <- mutate(metadata, Site = recode(.x=Site, "U of Michigan"="U Michigan"))
metadata <- mutate(metadata, Dx_Bin = recode(.x=Dx_Bin, "Cancer."="Cancer"))
metadata <- mutate(metadata, Gender = recode(.x=Gender, "f"="female", "m"="male"))
metadata <- rename_all(.tbl=metadata, .funs=tolower)
metadata <- rename(.data=metadata,
previous_history=hx_prev,
history_of_polyps=hx_of_polyps,
family_history_of_crc=hx_fam_crc,
diagnosis_bin=dx_bin,
diagnosis=dx,
sex=gender)
metadata <- mutate(metadata, diagnosis = factor(diagnosis, levels=c("normal", "adenoma", "cancer")))
dir.create("processed_data", showWarnings=FALSE)
write_tsv(x=metadata, path='processed_data/baxter.metadata.tsv')
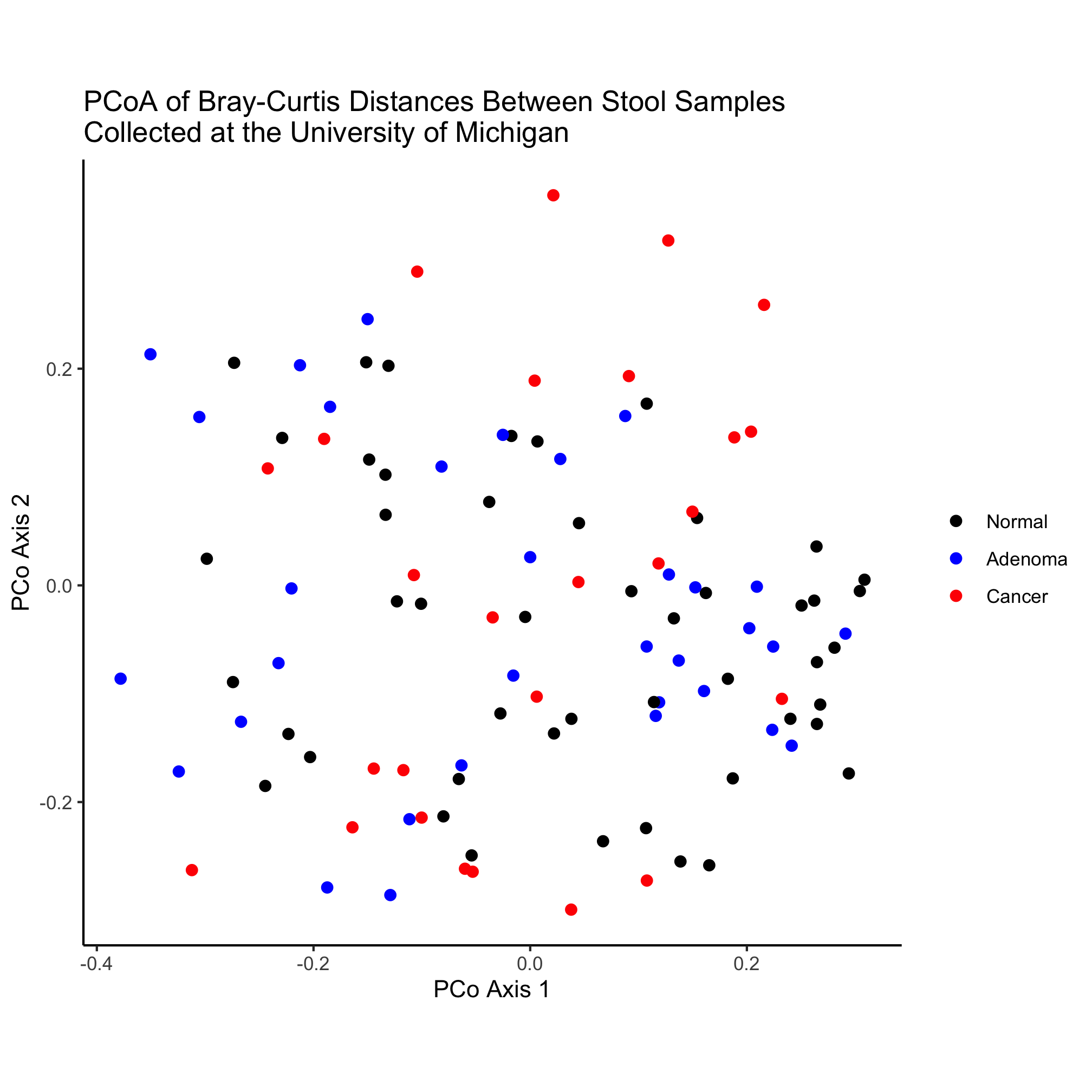
metadata_pcoa <- inner_join(metadata, pcoa, by=c('sample'='group'))
ggplot(metadata_pcoa, aes(x=axis1, y=axis2, color=diagnosis)) +
geom_point(shape=19, size=2) +
scale_color_manual(name=NULL,
values=c("black", "blue", "red"),
breaks=c("normal", "adenoma", "cancer"),
labels=c("Normal", "Adenoma", "Cancer")) +
coord_fixed() +
labs(title="PCoA of Bray-Curtis Distances Between Stool Samples",
x="PCo Axis 1",
y="PCo Axis 2") +
theme_classic()
We’ve almost accounted for all of the lines in this code chunk. One thing we haven’t accounted for yet is the inner_join function, which was called after cleaning up the metadata data frame and before using ggplot to plot the data. The syntax here should be somewhat clear. The function “joins” two data frames - pcoa and metadata - based on the columns “sample” and “group”, which are the columns in the metadata and pcoa data frames, respectively. If we were thinking ahead, we could have renamed the “group” column in pcoa to be “sample” and then the command could have just been inner_join(metadata, pcoa). When you look at the contents of the metadata_pcoa data frame you’ll see that the data frame is now 490 rows by 507 columns. Perhaps you’re wondering what the inner in the inner_join function is about. It turns out that the dplyr package has several ways to join data frames. As we’ll see, the inner_join function joins two data frames based on a column that they have in common (i.e. by=c('sample'='group') in our case) and if a sample or group is missing from one of the data frames, it is excluded from the joined data frame. This is what is called an “inner join”.
Joins
In addition to an “inner join”, the dplyr package has “left join” (i.e. left_join) and “right join” (i.e. right_join) functions, which will merge the data frames using the sample identifiers found in the left or right data frame being joined. There is also a “full join” (i.e. full_join), which produces a data frame where the samples from both data frames are represented even if they’re missing from one of the data frames. Let’s do a couple of examples to demonstrate these joins. To keep things simple, we’ll define two new data frames. We can do this by giving the tibble function a series of vectors that will be used to create the columns
a <- tibble(sample=c("A", "B", "C"), diagnosis=c("normal", "cancer", "adenoma"))
a
b <- tibble(sample=c("A", "B", "D"), previous_history=c(T, F, T))
b
## # A tibble: 3 x 2
## sample diagnosis
## <chr> <chr>
## 1 A normal
## 2 B cancer
## 3 C adenoma
## # A tibble: 3 x 2
## sample previous_history
## <chr> <lgl>
## 1 A TRUE
## 2 B FALSE
## 3 D TRUE
We’ll do a “left join” …
left_join(a, b, by="sample")
## # A tibble: 3 x 3
## sample diagnosis previous_history
## <chr> <chr> <lgl>
## 1 A normal TRUE
## 2 B cancer FALSE
## 3 C adenoma NA
Notice that because b doesn’t have a value for “C” in column “sample”, the resulting data frame has a NA in that cell. Because a doesn’t have a value for “D” in column “sample” it is excluded from the new data frame. If we instead do a “right join” …
right_join(a, b, by="sample")
## # A tibble: 3 x 3
## sample diagnosis previous_history
## <chr> <chr> <lgl>
## 1 A normal TRUE
## 2 B cancer FALSE
## 3 D <NA> TRUE
We see the opposite result - sample “C” is missing in the new data frame and the value in column “diagnosis” for sample “D” is NA. If we now do a “full join”…
full_join(a, b, by="sample")
## # A tibble: 4 x 3
## sample diagnosis previous_history
## <chr> <chr> <lgl>
## 1 A normal TRUE
## 2 B cancer FALSE
## 3 C adenoma NA
## 4 D <NA> TRUE
Here we see that all four samples are represented, but that the “diagnosis” and “previous_history” columns have NA values for samples D and C, respectively. Finally, returning to our old friend, “inner join”…
inner_join(a, b, by="sample")
## # A tibble: 2 x 3
## sample diagnosis previous_history
## <chr> <chr> <lgl>
## 1 A normal TRUE
## 2 B cancer FALSE
We now get a data frame that has two rows representing the two samples that were found in a and b. Depending on your goals, you will need to chose the appropriate join function. Most of the time I use an inner_join since I will only want the values (e.g. the axes in pcoa) that I have metadata for and I will only want the descriptors (e.g. the values in metadata) that I have community data for.
Activity 1
What happens in these cases when we reverse the a and b data frames in the inner_join function call?
Activity 2
What happens if we leave out the by="sample" argument from our join commands?
Perhaps we want to know whether there are any rows from our data frames that will be removed when we do an inner join. For this case, we can use the anti_join function from the dplyr package:
anti_join(a, b, by="sample")
## # A tibble: 1 x 2
## sample diagnosis
## <chr> <chr>
## 1 C adenoma
anti_join(b, a, by="sample")
## # A tibble: 1 x 2
## sample previous_history
## <chr> <lgl>
## 1 D TRUE
We can see that for the first case, the row for sample “C” is found in a, but not b. In the second case, sample “D” is found in b, but not a.
We can also see what from a overlaps with b and vice versa with the semi_join function from the dplyr package
semi_join(a, b, by="sample")
## # A tibble: 2 x 2
## sample diagnosis
## <chr> <chr>
## 1 A normal
## 2 B cancer
semi_join(b, a, by="sample")
## # A tibble: 2 x 2
## sample previous_history
## <chr> <lgl>
## 1 A TRUE
## 2 B FALSE
One last thing to comment on is that our simple examples of joining a and b have been using by="sample" in all of the examples. If you look at the syntax of the command we used when building our ordination plot, the syntax was by=c('sample'='group'). This is because sample is a column shared by both a and b, while sample and group are columns that contain the same information (i.e. the subject’s id number). Let’s illustrate this with a new data frame, c, which has a column group instead of sample:
c <- tibble(group=c("A", "B", "D"), previous_history=c(T, F, T))
c
## # A tibble: 3 x 2
## group previous_history
## <chr> <lgl>
## 1 A TRUE
## 2 B FALSE
## 3 D TRUE
If we do our inner_join as before, we’ll get an error…
inner_join(a, c, by="sample")
## Error: Join columns must be present in data.
## ✖ Problem with `sample`.
See that? It tells us that the join columns must be present in data and that there’s a problem wiht using “sample” to join the data frames. To resolve this, we need to use the syntax we saw earlier. We can replace by="sample" with by=c('sample'='group'). This effectively tells inner_join to join the two data frames using the sample column from data frame a and the group column from data frame b.
inner_join(a, c, by=c('sample'='group'))
## # A tibble: 2 x 3
## sample diagnosis previous_history
## <chr> <chr> <lgl>
## 1 A normal TRUE
## 2 B cancer FALSE
Selecting columns from our data frames
Looking at the ordination data that is in our pcoa data frame, we see that there were a few hundred columns. When this is joined to the metadata data frame we get a very wide and obnoxiously large data frame. We really only need the first four columns of the pcoa data frame (i.e. “group”, “axis1”, “axis2”, and “axis3”). We can do this with the select function from the dplyr package.
select(pcoa, group, axis1, axis2, axis3)
## # A tibble: 490 x 4
## group axis1 axis2 axis3
## <chr> <dbl> <dbl> <dbl>
## 1 2003650 0.281 -0.0575 -0.0517
## 2 2005650 0.309 0.00522 -0.0664
## 3 2007660 0.154 0.0622 -0.0178
## 4 2009650 0.293 -0.0153 0.0703
## 5 2013660 -0.223 -0.137 -0.0403
## 6 2015650 0.280 0.00598 -0.187
## 7 2017660 0.111 -0.0704 0.110
## 8 2019651 0.107 -0.224 0.0411
## 9 2023680 0.260 0.0965 -0.0790
## 10 2025653 -0.144 -0.169 0.0271
## # … with 480 more rows
The resulting tibble still has 490 rows, but now it has the 4 columns we selected. If we want to remove specific columns we could also use a negative sign
select(pcoa, -axis1)
## # A tibble: 490 x 490
## group axis2 axis3 axis4 axis5 axis6 axis7 axis8 axis9
## <chr> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2003… -0.0575 -0.0517 -0.0901 0.00955 -0.0599 -0.0529 -0.0391 0.0263
## 2 2005… 0.00522 -0.0664 0.0715 -0.00733 -0.0449 0.0400 -0.0223 -0.0268
## 3 2007… 0.0622 -0.0178 0.0953 -0.122 -0.0935 0.00763 0.0370 -0.0398
## 4 2009… -0.0153 0.0703 0.0351 -0.0176 -0.00318 0.0437 -0.0827 -0.0478
## 5 2013… -0.137 -0.0403 0.0344 -0.0115 -0.0400 0.107 0.233 0.0274
## 6 2015… 0.00598 -0.187 -0.0383 0.0653 -0.131 -0.164 0.0603 0.0522
## 7 2017… -0.0704 0.110 -0.104 0.0389 0.0439 0.0993 0.0709 -0.0981
## 8 2019… -0.224 0.0411 -0.103 -0.00765 0.0902 -0.107 0.00435 0.0413
## 9 2023… 0.0965 -0.0790 0.0406 -0.0554 -0.108 -0.0646 -0.0755 -0.0423
## 10 2025… -0.169 0.0271 0.108 -0.0600 -0.109 0.0316 -0.0612 -0.166
## # … with 480 more rows, and 481 more variables: axis10 <dbl>, axis11 <dbl>,
## # axis12 <dbl>, axis13 <dbl>, axis14 <dbl>, axis15 <dbl>, axis16 <dbl>,
## # axis17 <dbl>, axis18 <dbl>, axis19 <dbl>, axis20 <dbl>, axis21 <dbl>,
## # axis22 <dbl>, axis23 <dbl>, axis24 <dbl>, axis25 <dbl>, axis26 <dbl>,
## # axis27 <dbl>, axis28 <dbl>, axis29 <dbl>, axis30 <dbl>, axis31 <dbl>,
## # axis32 <dbl>, axis33 <dbl>, axis34 <dbl>, axis35 <dbl>, axis36 <dbl>,
## # axis37 <dbl>, axis38 <dbl>, axis39 <dbl>, axis40 <dbl>, axis41 <dbl>,
## # axis42 <dbl>, axis43 <dbl>, axis44 <dbl>, axis45 <dbl>, axis46 <dbl>,
## # axis47 <dbl>, axis48 <dbl>, axis49 <dbl>, axis50 <dbl>, axis51 <dbl>,
## # axis52 <dbl>, axis53 <dbl>, axis54 <dbl>, axis55 <dbl>, axis56 <dbl>,
## # axis57 <dbl>, axis58 <dbl>, axis59 <dbl>, axis60 <dbl>, axis61 <dbl>,
## # axis62 <dbl>, axis63 <dbl>, axis64 <dbl>, axis65 <dbl>, axis66 <dbl>,
## # axis67 <dbl>, axis68 <dbl>, axis69 <dbl>, axis70 <dbl>, axis71 <dbl>,
## # axis72 <dbl>, axis73 <dbl>, axis74 <dbl>, axis75 <dbl>, axis76 <dbl>,
## # axis77 <dbl>, axis78 <dbl>, axis79 <dbl>, axis80 <dbl>, axis81 <dbl>,
## # axis82 <dbl>, axis83 <dbl>, axis84 <dbl>, axis85 <dbl>, axis86 <dbl>,
## # axis87 <dbl>, axis88 <dbl>, axis89 <dbl>, axis90 <dbl>, axis91 <dbl>,
## # axis92 <dbl>, axis93 <dbl>, axis94 <dbl>, axis95 <dbl>, axis96 <dbl>,
## # axis97 <dbl>, axis98 <dbl>, axis99 <dbl>, axis100 <dbl>, axis101 <dbl>,
## # axis102 <dbl>, axis103 <dbl>, axis104 <dbl>, axis105 <dbl>, axis106 <dbl>,
## # axis107 <dbl>, axis108 <dbl>, axis109 <dbl>, …
The result is that the “axis1” column has been removed. If we consider our metadata data frame, we could also select the sample column any column that starts with “diagnosis”
select(metadata, sample, starts_with("diagnosis"))
## # A tibble: 490 x 3
## sample diagnosis_bin diagnosis
## <chr> <chr> <fct>
## 1 2003650 High Risk Normal normal
## 2 2005650 High Risk Normal normal
## 3 2007660 High Risk Normal normal
## 4 2009650 Adenoma adenoma
## 5 2013660 Normal normal
## 6 2015650 High Risk Normal normal
## 7 2017660 Cancer cancer
## 8 2019651 Normal normal
## 9 2023680 High Risk Normal normal
## 10 2025653 Cancer cancer
## # … with 480 more rows
This gets us a new data frame with the columns “sample”, “diagnosis_bin”, and “diagnosis”. We could also get the “sample” column and any column that contains “history”
select(metadata, sample, contains("history"))
## # A tibble: 490 x 4
## sample previous_history history_of_polyps family_history_of_crc
## <chr> <lgl> <lgl> <lgl>
## 1 2003650 FALSE TRUE TRUE
## 2 2005650 FALSE TRUE FALSE
## 3 2007660 FALSE TRUE TRUE
## 4 2009650 FALSE TRUE FALSE
## 5 2013660 FALSE FALSE FALSE
## 6 2015650 FALSE TRUE FALSE
## 7 2017660 TRUE TRUE FALSE
## 8 2019651 FALSE FALSE FALSE
## 9 2023680 TRUE TRUE FALSE
## 10 2025653 TRUE TRUE FALSE
## # … with 480 more rows
This generates a data frame that contains the columns “sample”, “previous_history”, “history_of_polyps”, and “family_history_of_crc”. There are other helper functions including ends_with, matches, num_range, and one_of that you can learn more about by using the ? helper.
Selecting rows from our data frames
We might also want to make new data frames that contain a subset of the rows. We can “filter” the data frame using the filter function from the dplyr package. Let’s assume that we want to recreate our favorite ordination using only samples from the University of Michigan. We can generate a new data frame using filter
filter(metadata, site=="U Michigan")
## # A tibble: 107 x 17
## sample fit_result site diagnosis_bin diagnosis previous_history
## <chr> <dbl> <chr> <chr> <fct> <lgl>
## 1 20036… 0 U Mi… High Risk No… normal FALSE
## 2 20056… 0 U Mi… High Risk No… normal FALSE
## 3 20076… 26 U Mi… High Risk No… normal FALSE
## 4 20136… 0 U Mi… Normal normal FALSE
## 5 20196… 19 U Mi… Normal normal FALSE
## 6 20256… 1509 U Mi… Cancer cancer TRUE
## 7 20296… 0 U Mi… Adenoma adenoma FALSE
## 8 20416… 0 U Mi… Adenoma adenoma FALSE
## 9 20456… 0 U Mi… Normal normal FALSE
## 10 20576… 0 U Mi… High Risk No… normal FALSE
## # … with 97 more rows, and 11 more variables: history_of_polyps <lgl>,
## # age <dbl>, sex <chr>, smoke <lgl>, diabetic <lgl>,
## # family_history_of_crc <lgl>, height <dbl>, weight <dbl>, nsaid <lgl>,
## # diabetes_med <lgl>, stage <chr>
The resulting data frame has 107 samples. You’ll notice that I used site=="U Michigan". This tells filter to identify those rows where the “site” column had a value equal to “U Michigan”. The == is a logical comparison that asks whether the value on either side of the == are the same. The answer is either TRUE or FALSE. There are other logical operators that you should already be familiar with (but perhaps didn’t know!) including <, <=, >, >=. These should be self explanatory. The fit result measures how much blood is in a person’s stool. It’s a common non-invasive diagnostic to identify colonic lesions and a value greater than 100 is a concern. If we want ever subject that has a fit_result greater than or equal to 100 we would write
filter(metadata, fit_result >= 100)
## # A tibble: 126 x 17
## sample fit_result site diagnosis_bin diagnosis previous_history
## <chr> <dbl> <chr> <chr> <fct> <lgl>
## 1 20256… 1509 U Mi… Cancer cancer TRUE
## 2 20936… 286 Dana… Normal normal TRUE
## 3 21056… 314 U Mi… Normal normal FALSE
## 4 21856… 982 Toro… Adv Adenoma adenoma FALSE
## 5 21876… 1200 Toro… Cancer cancer FALSE
## 6 22036… 1992 U Mi… Cancer cancer TRUE
## 7 22556… 140 Dana… Cancer cancer TRUE
## 8 22676… 149 Dana… Cancer cancer TRUE
## 9 22836… 1346 Toro… Cancer cancer FALSE
## 10 22876… 939 Dana… Cancer cancer TRUE
## # … with 116 more rows, and 11 more variables: history_of_polyps <lgl>,
## # age <dbl>, sex <chr>, smoke <lgl>, diabetic <lgl>,
## # family_history_of_crc <lgl>, height <dbl>, weight <dbl>, nsaid <lgl>,
## # diabetes_med <lgl>, stage <chr>
Some of our columns are already logical. To get those individuals with a previous history of colorectal cancer we could do
filter(metadata, previous_history)
## # A tibble: 138 x 17
## sample fit_result site diagnosis_bin diagnosis previous_history
## <chr> <dbl> <chr> <chr> <fct> <lgl>
## 1 20176… 7 Dana… Cancer cancer TRUE
## 2 20236… 0 Dana… High Risk No… normal TRUE
## 3 20256… 1509 U Mi… Cancer cancer TRUE
## 4 20336… 0 Toro… High Risk No… normal TRUE
## 5 20516… 0 Dana… Adenoma adenoma TRUE
## 6 20556… 0 Dana… Adv Adenoma adenoma TRUE
## 7 20636… 0 Dana… High Risk No… normal TRUE
## 8 20876… 5 Dana… High Risk No… normal TRUE
## 9 20936… 286 Dana… Normal normal TRUE
## 10 21096… 0 Dana… Normal normal TRUE
## # … with 128 more rows, and 11 more variables: history_of_polyps <lgl>,
## # age <dbl>, sex <chr>, smoke <lgl>, diabetic <lgl>,
## # family_history_of_crc <lgl>, height <dbl>, weight <dbl>, nsaid <lgl>,
## # diabetes_med <lgl>, stage <chr>
If we want those samples from people without a previous history we can use the ! operator which turns TRUE to FALSE and FALSE to TRUE
filter(metadata, !previous_history)
## # A tibble: 349 x 17
## sample fit_result site diagnosis_bin diagnosis previous_history
## <chr> <dbl> <chr> <chr> <fct> <lgl>
## 1 20036… 0 U Mi… High Risk No… normal FALSE
## 2 20056… 0 U Mi… High Risk No… normal FALSE
## 3 20076… 26 U Mi… High Risk No… normal FALSE
## 4 20096… 10 Toro… Adenoma adenoma FALSE
## 5 20136… 0 U Mi… Normal normal FALSE
## 6 20156… 0 Dana… High Risk No… normal FALSE
## 7 20196… 19 U Mi… Normal normal FALSE
## 8 20276… 0 Toro… Normal normal FALSE
## 9 20296… 0 U Mi… Adenoma adenoma FALSE
## 10 20316… 0 Toro… Adenoma adenoma FALSE
## # … with 339 more rows, and 11 more variables: history_of_polyps <lgl>,
## # age <dbl>, sex <chr>, smoke <lgl>, diabetic <lgl>,
## # family_history_of_crc <lgl>, height <dbl>, weight <dbl>, nsaid <lgl>,
## # diabetes_med <lgl>, stage <chr>
The ! can also be used as != to test whether two values are different from each other. We could use this to get the samples from people that do not have a normal diagnosis
filter(metadata, diagnosis != 'normal')
## # A tibble: 318 x 17
## sample fit_result site diagnosis_bin diagnosis previous_history
## <chr> <dbl> <chr> <chr> <fct> <lgl>
## 1 20096… 10 Toro… Adenoma adenoma FALSE
## 2 20176… 7 Dana… Cancer cancer TRUE
## 3 20256… 1509 U Mi… Cancer cancer TRUE
## 4 20296… 0 U Mi… Adenoma adenoma FALSE
## 5 20316… 0 Toro… Adenoma adenoma FALSE
## 6 20356… 0 Toro… Adv Adenoma adenoma FALSE
## 7 20376… 72 Toro… Cancer cancer FALSE
## 8 20416… 0 U Mi… Adenoma adenoma FALSE
## 9 20496… 0 Dana… Adenoma adenoma FALSE
## 10 20516… 0 Dana… Adenoma adenoma TRUE
## # … with 308 more rows, and 11 more variables: history_of_polyps <lgl>,
## # age <dbl>, sex <chr>, smoke <lgl>, diabetic <lgl>,
## # family_history_of_crc <lgl>, height <dbl>, weight <dbl>, nsaid <lgl>,
## # diabetes_med <lgl>, stage <chr>
Activity 3
A common bug for novice and experienced programmers is shown below
filter(metadata, diagnosis="cancer")
Can you see what the problem is and how to fix it?
Activity 4
Create a data frame that contains only females
Activity 5
Create a data frame that contains individuals are 50 years old and younger
Activity 6
Use the filter command to generate an ordination of samples from the University of Michigan.
The filter and select functions are very powerful for subsetting our data frames. What if I want to get those samples from people that have a fit result over 100 and were given a normal diagnosis? We can use the & operator to see if two logical comparisons are true
filter(metadata, fit_result >= 100 & diagnosis == "normal")
## # A tibble: 5 x 17
## sample fit_result site diagnosis_bin diagnosis previous_history
## <chr> <dbl> <chr> <chr> <fct> <lgl>
## 1 20936… 286 Dana… Normal normal TRUE
## 2 21056… 314 U Mi… Normal normal FALSE
## 3 23216… 148 Dana… Normal normal FALSE
## 4 30996… 356 Dana… High Risk No… normal TRUE
## 5 31376… 118 U Mi… Normal normal FALSE
## # … with 11 more variables: history_of_polyps <lgl>, age <dbl>, sex <chr>,
## # smoke <lgl>, diabetic <lgl>, family_history_of_crc <lgl>, height <dbl>,
## # weight <dbl>, nsaid <lgl>, diabetes_med <lgl>, stage <chr>
If we want samples from people with a high fit result or a cancer diagnosis we can use a similar approach, except that instead of using & we would use |
filter(metadata, fit_result >= 100 | diagnosis == "cancer")
## # A tibble: 156 x 17
## sample fit_result site diagnosis_bin diagnosis previous_history
## <chr> <dbl> <chr> <chr> <fct> <lgl>
## 1 20176… 7 Dana… Cancer cancer TRUE
## 2 20256… 1509 U Mi… Cancer cancer TRUE
## 3 20376… 72 Toro… Cancer cancer FALSE
## 4 20936… 286 Dana… Normal normal TRUE
## 5 21056… 314 U Mi… Normal normal FALSE
## 6 21856… 982 Toro… Adv Adenoma adenoma FALSE
## 7 21876… 1200 Toro… Cancer cancer FALSE
## 8 22036… 1992 U Mi… Cancer cancer TRUE
## 9 22556… 140 Dana… Cancer cancer TRUE
## 10 22676… 149 Dana… Cancer cancer TRUE
## # … with 146 more rows, and 11 more variables: history_of_polyps <lgl>,
## # age <dbl>, sex <chr>, smoke <lgl>, diabetic <lgl>,
## # family_history_of_crc <lgl>, height <dbl>, weight <dbl>, nsaid <lgl>,
## # diabetes_med <lgl>, stage <chr>
We can make complicated filter commands easier to understand by grouping the logical questions with parentheses. Because algebra within parentheses is evaluated first, we can control the order of operations for our question. For example, if we want the samples from women who have a high fit result or a cancer diagnosis we might be tempted do do…
filter(metadata, fit_result >= 100 | diagnosis == "cancer" & sex == "female") %>%
count(sex)
## # A tibble: 2 x 2
## sex n
## <chr> <int>
## 1 female 67
## 2 male 76
What happened? If you look through a table that lists the order of operations, you’ll notice that the logical AND is perormed before the logical OR. Who remembers such things? To make our intention more clear and get the correct answer, we can wrap the OR statement in parentheses
filter(metadata, (fit_result >= 100 | diagnosis == "cancer") & sex == "female") %>%
count(sex)
## # A tibble: 1 x 2
## sex n
## <chr> <int>
## 1 female 67
Activity 7
Create a data frame that contains samples from individuals who are 50 years old and younger and have a non-normal diagnosis
Activity 8
Create a data frame that contains samples from individuals who have a previous or family history of colorectal cancer
Selecting and filtering with pipes
Let’s leverage the select and filter commands we have been using to work with a new tsv file. The file raw_data/baxter.groups.ave-std.summary was generated by the mothur summary.single command, which rarefies the number of sequences per sample and calculates a variety of alpha diversity metrics.
Activity 9
This file has a number of columns that aren’t that interesting for us. You will also find that the method column has two values - ave and std - which indicate the average value of the alpha diversity metric after rarefying and the standard deviation (i.e. std), which is the standard deviation for the rarefaction replicates. You have several tasks…
- Write the code needed to read in the file to a new data frame called
alpha. Make sure that the group column is read in as characters - Filter out the rows that contain the standard deviation data
- Select the columns that contain the subject identifier, the number of observed OTUs (i.e “sobs”), the Shannon diversity index (i.e. “shannon”), the inverse Simpson diversity index (i.e. “invsimpson”), and Good’s coverage (i.e. “coverage”)
- Use the appropriate join function to create a data frame called
meta_alphathat is a join betweenmetadataandalpha
Hopefully that was a good review of what we’ve done in this and the previous lessons. The approach we’ve taken to generate meta_alpha works perfectly. I’d like to show you a different way to think about the code. If you look at these four lines of code, you should see that the data kind of “flows” from the tsv file to the final version of alpha before we join it to metadata. There’s a package installed with dplyr called magrittr that has a funny looking function called a pipe - %>%. The pipe, directs the flow of data from one command to the next. Instead of writing over alpha multiple times, we can write it once as the output of the data flow through the pipes.
alpha <- read_tsv(file="raw_data/baxter.groups.ave-std.summary", col_types=cols(group = col_character())) %>%
filter(method=='ave') %>%
select(group, sobs, shannon, invsimpson, coverage)
alpha
## # A tibble: 490 x 5
## group sobs shannon invsimpson coverage
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 2003650 262. 4.02 35.6 0.991
## 2 2005650 292. 3.98 26.6 0.990
## 3 2007660 283. 3.91 26.7 0.992
## 4 2009650 324. 4.16 30.1 0.991
## 5 2013660 133. 3.33 17.4 0.997
## 6 2015650 233. 3.74 20.8 0.993
## 7 2017660 238. 3.98 28.6 0.994
## 8 2019651 191. 3.69 20.1 0.996
## 9 2023680 300. 4.01 28.9 0.992
## 10 2025653 179. 3.39 14.5 0.995
## # … with 480 more rows
Viola! Cool, eh? You may not see the benefit of the pipes here, but in subsequent lessons we will pipe together numerous functions to direct the flow of data. Instead of writing over alpha as we did in the previous code chunks, some people would rather write each update to a new variable name. Both approaches get tedious and so the ability to pipe becomes pretty handy. In fact, we can skip the creation of the alpha data frame all together by piping this flow right into the inner_join function call. Notice that in the code below, the inner_join function call has a . where alpha had been before. The . tells inner_join to use the data that is flowing through the pipe.
meta_alpha <- read_tsv(file="raw_data/baxter.groups.ave-std.summary", col_types=cols(group = col_character())) %>%
filter(method=='ave') %>%
select(group, sobs, shannon, invsimpson, coverage) %>%
inner_join(metadata, ., by=c("sample"="group"))
meta_alpha
## # A tibble: 490 x 21
## sample fit_result site diagnosis_bin diagnosis previous_history
## <chr> <dbl> <chr> <chr> <fct> <lgl>
## 1 20036… 0 U Mi… High Risk No… normal FALSE
## 2 20056… 0 U Mi… High Risk No… normal FALSE
## 3 20076… 26 U Mi… High Risk No… normal FALSE
## 4 20096… 10 Toro… Adenoma adenoma FALSE
## 5 20136… 0 U Mi… Normal normal FALSE
## 6 20156… 0 Dana… High Risk No… normal FALSE
## 7 20176… 7 Dana… Cancer cancer TRUE
## 8 20196… 19 U Mi… Normal normal FALSE
## 9 20236… 0 Dana… High Risk No… normal TRUE
## 10 20256… 1509 U Mi… Cancer cancer TRUE
## # … with 480 more rows, and 15 more variables: history_of_polyps <lgl>,
## # age <dbl>, sex <chr>, smoke <lgl>, diabetic <lgl>,
## # family_history_of_crc <lgl>, height <dbl>, weight <dbl>, nsaid <lgl>,
## # diabetes_med <lgl>, stage <chr>, sobs <dbl>, shannon <dbl>,
## # invsimpson <dbl>, coverage <dbl>
But wait… there’s more!
read_tsv(file="raw_data/baxter.groups.ave-std.summary", col_types=cols(group = col_character())) %>%
filter(method=='ave') %>%
select(group, sobs, shannon, invsimpson, coverage) %>%
inner_join(metadata, ., by=c("sample"="group")) %>%
ggplot(aes(x=age, y=shannon, color=diagnosis)) +
geom_point(shape=19, size=2) +
coord_cartesian(xlim=c(0,90), ylim=c(0,5)) +
scale_color_manual(name=NULL,
values=c("black", "blue", "red"),
breaks=c("normal", "adenoma", "cancer"),
labels=c("Normal", "Adenoma", "Cancer")) +
labs(title="Relationship between community diversity and subject's age",
x="Age",
y="Shannon Diversity Index") +
theme_classic()
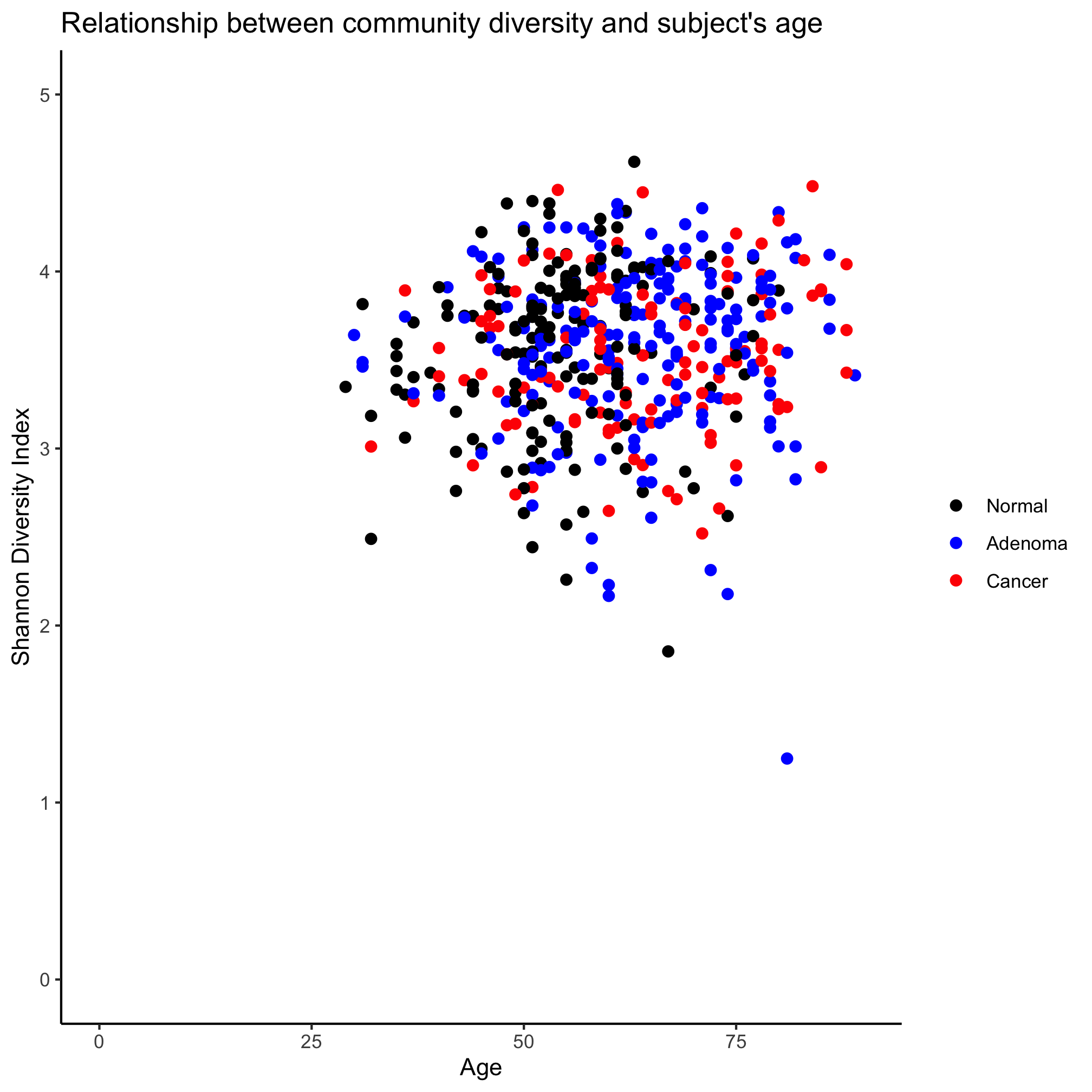
We’ve gone all the way - reading in the data from a tsv file to getting the rows and columns we want to joining it with our metadata to plotting. All in one command. Pretty slick.
Activity 10
With our new found piping skillz, rewrite the code from the end of the last tutorial to generate the ordination. Use the metadata data frame that we’ve already been working with
Conclusion
A couple of closing thoughts are needed before we move on to the next lesson where we’ll start doing more sophisticated work with data frames and functions from the dplyr package. First, you might ask why we ran select and filter on alpha rather than on the output of the inner_join. There’s no real reason. The output would be the same. Do what makes sense for where you are in your analysis. Second, you can feel free to break up the piping as much as you want. It is there as a helper to your coding so that you don’t have to create temporary data frames or write over ones you just made. Beyond these advantages, most people find that debugging code that uses the pipes is much easier than with the other approaches. Finally, instead of making the alpha diversity scatter plot one pipeline, I probably would normally break it up into two pipelines. One pipeline to create alpha and do the select and filter steps. The second would join alpha with metadata and produce the plot.